Method Article
CRISPR Epigenome Editing in Human Cells using Plasmid DNA Transfection and mRNA Nucleofection Delivery
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
The protocol describes methods for Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based epigenome editing in human cell lines using plasmid DNA transfection and mRNA nucleofection.
Streszczenie
Epigenetics refers to chemical modifications of histone proteins and DNA that can regulate the expression of genes. The human epigenome is altered dynamically during cell differentiation and aging, and many diseases are associated with aberrant epigenome patterning. Recent advances in CRISPR have led to the development of programmable tools to edit epigenetic modifications at targeted genomic loci, enabling precise rewriting of epigenetic modifications in human cells. CRISPR-based epigenome editors rely on catalytically dead Cas9 coupled with epigenetic modifiers that ultimately result in programmed repression or activation of targeted genes in mammalian genomes. Unlike traditional genome editing methods, epigenome editing does not require DNA breaks or changes in the human genome sequence and thus serves as a safer alternative to control gene expression. In this protocol, we highlight two different methods to perform dCas9-mediated epigenome editing in human cell lines using plasmid DNA transfections and nucleofection of mRNAs encoding CRISPR epigenome editors. We demonstrate programmable epigenome editing to transiently repress genes using CRISPR interference (CRISPRi) and for silencing genes durably for many weeks using CRISPRoff, a fusion of dCas9 to the KRAB domain and de novo DNA methyltransferase complex. We also provide guidance on quantitative methods to measure successful epigenome editing of target genes and key considerations on which epigenome editing tool to use, depending on experimental criteria.
Wprowadzenie
Though the genomic content of every cell in our body is nearly identical, the transcriptional profile of each cell type differs greatly. Epigenetic modifications on DNA and histone proteins are key regulators of transcriptional expression. Transcriptionally active euchromatin is hallmarked by distinct epigenetic marks compared to compact, transcriptionally inactive heterochromatin. For example, heterochromatic regions are defined by repressive histone modifications, including trimethylation on lysine 9 of histone 3 (H3K9me3), trimethylation on lysine 27 of histone H3 (H3K27me3), and DNA methylation on cytosines next to guanines (CpG) at gene promoters1. Genomic regions of active gene expression are defined typically by histone acetylation and trimethylation on lysine 4 of histone 3 (H3K4me3)1.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) revolution has generated a wealth of tools that enable programmed altering of genomic sequences. CRISPR technology is based on a prokaryotic defense mechanism capable of cleaving nucleic acids at programmable target sequences. CRISPR nucleases2,3,4, base editors5,6, and prime editors7,8 can change the DNA sequence of mammalian genomes through DNA cutting and repair of these breaks. Though effective, these strategies can cause DNA breaks at off-target sites9,10 and large-scale genomic structural mutations11,12,13,14,15. Alternative CRISPR-based tools enable tractable modulation of gene activation and repression without changing the underlying DNA sequence. These tools harness a nuclease deficient Cas9 (dCas9), allowing for DNA binding at target sites dictated by sgRNA sequence, in combination with effector proteins that alter the chromatin landscape16,17,18. Effector proteins, such as epigenetic writers, readers, and erasers, can be directly fused to dCas9 or recruited by a peptide scaffold fused to dCas9, such as SunTag, or an RNA scaffold on the sgRNA, such as the MS2-MCP system16,17,18. Examples of programmable transcription control tools include CRISPR activation (CRISPRa)19,20,21 and CRISPR interference (CRISPRi)22,23. CRISPRa functions by directly recruiting the transcription machinery, increasing target transcriptional gene expression19. In contrast, CRISPRi represses transcription by establishing H3K9me3, a repressive epigenetic mark22.
Advances in epigenome editing have enabled broad use of these tools across scientific fields. Fusions of different effector domains and proteins have expanded the toolkit of available epigenome editors18,24,25,26,27,28,29,30. Additionally, epigenome editors are used to decipher the roles of epigenetic modifications31,32,33,34,35,36, effectors37,38,39, and effector mutations28,40,41 in gene regulation. Specifically, CRISPRi and CRISPRa are used in functional genomics screens for a variety of biological processes, including cell survival42 and cell fate43,44,45,46,47. Furthermore, epigenome editing holds therapeutic potential for ex vivo cell engineering and in vivo therapies18.
Here, we describe methods for applying two dCas9-based epigenome editors for programmable transcriptional repression in human cell lines: CRISPRi22 and CRISPRoff48. CRISPRi is a fusion of dCas9 to the repressive KRAB domain from a zinc finger protein such as ZNF10 (KOX1) and ZIM322,23. When CRISPRi is targeted to a specific gene promoter, the KRAB domain recruits an H3K9me3 writer, SETDB1, to repress the target gene (Figure 1A). When CRISPRi is expressed transiently, the established H3K9me3 at the target locus is not maintained, and gene expression is restored over time32,48. To conduct stable knockdowns using CRISPRi such as in functional genomics applications, constitutive expression of CRISPRi in the cells along with the sgRNA is essential. Recently, CRISPRoff was engineered to program heritable epigenome editing48. CRISPRoff is a single protein fusion of dCas9 to the KRAB domain and de novo DNA methyltransferase complex, DNMT3A and DNMT3L. A transient pulse of CRISPRoff in human cells programs deposition of H3K9me3 and DNA methylation at the targeted genes, which leads to long-term repression of target genes by the maintenance of DNA methylation and H3K9me3 (Figure 1B)48. Furthermore, epigenome edits can be reversed. For example, a gene that is stably silenced by CRISPRoff can be reactivated by TET1-dCas9 which can enzymatically remove the DNA methylation marks at target loci49.
This protocol will detail two delivery methods for the transient expression of epigenome editors: plasmid DNA transfection and mRNA nucleofection. Additionally, we outline how to use flow cytometry to assess epigenome editing efficacy at two endogenous genes, CLTA and CD55. These methods can be adapted and applied to other epigenome editing experiments using additional editors or can be used for targeting different genes.

Figure 1: Schematic of CRISPRi and CRISPRoff epigenome editing mechanism and workflow. (A) Linear schematics of the sgRNA transgene and CRISPRi epigenome editor. The addition of CRISPRi and sgRNA allows for the addition of the repressive H3K9me3 histone mark to silence the target locus. The highest level of silencing is achieved early after CRISPRi addition, and the gene target is generally reactivated after a few passages. (B) Linear schematics of the sgRNA transgene and CRISPRoff epigenome editor. The addition of CRISPRoff to cells targeting a gene of interest leads to the addition of repressive H3K9me3 along with DNA methylation at CpG sites to silence the target gene. Silencing by CRISPRoff is heritable - the high level of silencing is achieved early during transfection and persists over multiple cell divisions. (C) Overview of timeline for epigenome editing via transfection method. On day 0, the cells are plated for transfection. On day 1, the editor and guide plasmid can be introduced into the cells via transfection. On day 3, the cells will be assessed for BFP expression via flow cytometry. The percent of BFP is used as a normalizing factor to determine the final silencing efficacy of the experiment for each condition. On day 6 onwards, the cells are analyzed for silencing the reporter of interest as the highest level of silencing will be achieved on this day. (D) Transfection method overview wherein epigenome editor and sgRNA plasmids are added to cells in a dropwise manner. (E) Overview of the timeline for epigenome editing via the nucleofection methods. In this protocol, mRNA is nucleofection into the cells on day 0. The cells are assessed for silencing on day 3 post-nucleofection using flow cytometry analysis. (F) Overview of the nucleofection protocol. Appropriate amounts of cells and mRNA are mixed and added into nucleofector cuvettes. If the sgRNA is introduced by nucleofection, it can also be added to this mixture. The cuvettes are put into the nucleofector, and appropriate pulse codes are used to introduce the mRNA into cells. Post-nucleofection, the cells are plated and passaged for analysis in later days. (G) Comparison between plasmid transfection and mRNA nucleofection strategies for epigenome editing. Please click here to view a larger version of this figure.
Protokół
NOTE: Supplementary File 1 contains details about the sgRNA design, cloning, and cell line generation of our representative data. The representative results section also details suggestions for controls.
1. Transfection of epigenome editor-expressing plasmids into HEK293T cells
NOTE: This protocol describes the delivery of CRISPR-encoding plasmids into HEK293T cells. We have engineered cells to express a sgRNA (Addgene 217306) targeting the promoter region of CLTA, a non-essential gene, which is endogenously tagged with GFP. The CLTA-GFP HEK 293T cells originated from a previous study50. In this example, the epigenome editors are fused directly to a blue fluorescent protein (BFP), which allows us to quantify transfection efficiency and ensures accurate evaluation of experimental conditions. The efficacy of the approach is demonstrated by the silencing of CLTA-GFP, which can be measured quantitatively at the protein level in single cells using flow cytometry.
- Cell culture
- Maintain HEK293T cells in high glucose DMEM with 10% FBS and 1x Penicillin-Streptomycin-Glutamine. Passage the cells every 2-3 days, ensuring they remain at a confluency of 60%-70%.
NOTE: To ensure optimal transfection efficiency, it is critical to maintain cell confluency below 70%, which can be monitored daily under a microscope.
- Maintain HEK293T cells in high glucose DMEM with 10% FBS and 1x Penicillin-Streptomycin-Glutamine. Passage the cells every 2-3 days, ensuring they remain at a confluency of 60%-70%.
- Day 0: Seeding HEK293T cells for transfection
- The day before transfection, count cells with an automated cell counter using trypan blue live/dead staining and seed ~1.5 x 104 live cells per well in a 96-well plate. Keep the final volume of each well at 200 µL. Cells should reach 60%-70% confluency the next day.
NOTE: This can also be performed in 24-well or 6-well plates efficiently. See Table 1 for cell numbers and DNA amount for transfection.
- The day before transfection, count cells with an automated cell counter using trypan blue live/dead staining and seed ~1.5 x 104 live cells per well in a 96-well plate. Keep the final volume of each well at 200 µL. Cells should reach 60%-70% confluency the next day.
- Day 1: Transfection of CRISPRoff and CRISPRi plasmids into HEK29T cells
- Aliquot 150 ng of the epigenome editor plasmid (Addgene 167981) for each well in 3 µL total volume. These plasmid dilutions can be prepared in PCR strip tubes.
NOTE: If epigenome editor and the sgRNA plasmid are being co-transfected, then we recommend trying a 3:4 molar ratio of editor:sgRNA plasmids. The ratio may need to be optimized to yield the highest efficiency of epigenome editing. - Prewarm the transfection reagent and minimal essential medium to room temperature about 30-60 min prior to transfection. Mix 0.6 µL of transfection reagent and 20 µL of minimal essential medium per well and incubate at room temperature for 15 min.
NOTE: A master mix of transfection reagent and minimal essential medium can be made if performing multiple transfections. - Add 20.6 µL of the transfection reagent and minimal essential medium mixture to each PCR strip tube containing 3 µL of plasmid DNA. Allow to sit at room temperature for 15 min.
- Add mixture slowly to the plate of seeded cells and tap the plate gently to disperse the mixture. Include an un-transfected control to provide a proper baseline for comparison in downstream analyses.
NOTE: When adding the transfection mixture to seeded cells, it is essential to do so dropwise and gently to minimize disturbance, as this can adversely affect transfection efficiency.
- Aliquot 150 ng of the epigenome editor plasmid (Addgene 167981) for each well in 3 µL total volume. These plasmid dilutions can be prepared in PCR strip tubes.
- Day 2: Check transfection success
- Examine cells under a microscope at 10X magnification to determine if they are BFP positive, as this indicates successful transfection. Ensure the cells are not disturbed during the observation process.
- Day 3: Split the 96-well plate and assess transfection efficiency using the BFP marker
NOTE: A successful transfection should yield at least 30% of cells as BFP positive. However, with dCas9, CRISPRi, and CRISPRoff, we typically detect more than 60% BFP positive cells two days after transfection. Transfection efficiency is negatively correlated with plasmid size, so different epigenome editors may yield different transfection efficiencies.- Prewarm trypsin and DMEM with 10% FBS and 1x Penicillin-Streptomycin-Glutamine to room temperature for about 30-60 min prior to use.
- Aspirate media from the wells; be careful not to remove cells attached to the bottom. Add 50 µL of trypsin per well. Incubate for 2-3 min at 37 °C.
NOTE: It is not unusual to observe some floating cells, especially following transfection with CRISPRoff. We hypothesize this cell death is due to toxicity of DNMT3A-3L overexpression. - Inhibit trypsin by adding 150 µL of complete DMEM per well. Pipette up and down to resuspend cells and split cells for confluency on day 5 (~20 µL of cell mixture in 200 µL of media of a 96-well plate).
- Use 50 µL of resuspended cells for flow cytometry to assess transfection efficiency.
2. Nucleofection of epigenome editor mRNA into K-562 cells
NOTE: This section details the process of nucleofecting CRISPRoff mRNA into K-562 cells. For simplicity, we have pre-engineered the K-562 cells to constitutively express a sgRNA that targets the promoter of the CD55 endogenous gene (Addgene 217306). Delivering CRISPRoff mRNA directly to the cells has the potential to decrease the cellular toxicity that accompanies plasmid DNA-based approaches while still achieving similar gene silencing efficacy. Additionally, nucleofection can be used to introduce epigenome editor constructs in cell lines that are challenging to transfect efficiently, such as K-562s.
- Cell culture
- Prior to nucleofection, maintain K-562 cells in a flask with RPMI with 10% FBS and 1x Penicillin-Streptomycin-Glutamine. Passage the cells every day, ensuring that they remain at a confluency of 60%-70%.
- On the day of nucleofection, thaw and gently vortex CRISPRoff mRNA in a sterile RNase-free microcentrifuge tube and store the mRNA on ice. Use 2 µg of mRNA per 2.0 x 105 cells in each well of a strip nucleocuvette. Ensure that the volume of mRNA does not exceed 10% of the total nucleofection volume (22 µL).
NOTE: Nucleofection can also be performed in larger cuvettes with more cells and mRNA. We suggest scaling the amount of mRNA proportionally to the number of cells you will be adding. For example, when performing a nucleofection in a large cuvette on 1 x 106 cells, we suggest using 10 µg of mRNA, with a total nucleofection volume of 100 µL. Additionally, the dosage of the mRNA can be optimized to ensure high editing efficacy. We often use 2-5 µg of editor mRNA for every 2.0 x 105 cells for efficient results. Free nucleotides that were not incorporated during mRNA synthesis should be removed before quantification. - Prepare the nucleofection solution according to the manufacturer's instructions and warm to room temperature for 15 min before nucleofection.
NOTE: The nucleofector solution may be different or need to be optimized for different cell types. - Harvest and count K-562 cells using an automated cell counter with trypan blue live/dead staining. For nucleofection in a strip cuvette, aliquot ~2.0 x 105 cells per sample into a sterile microcentrifuge tube.
- Centrifuge the cells at 500 x g for 5 min at room temperature, then discard the supernatant. Wash the cells 1x with room temperature PBS at 500 x g for 5 min, then discard the supernatant.
- Calculate the amount of nucleofector solution to resuspend cells in by subtracting the mRNA volume from 22 µL. Resuspend the cells in the appropriate amount of nucleofector solution.
- Add cell solution to 2 µg of CRISPRoff mRNA (volume determined in step 2.1.2) and transfer the solution to a cuvette, being careful not to form bubbles, as this may compromise the efficiency of nucleofection. Gently tap the nucleocuvette to ensure that cells are at the bottom.
- Nucleofect the cells using the 4D-Nucleofector system with the appropriate pulse code. The FF-120 code is recommended for K-562 cells.
NOTE: The pulse code may need to be optimized for different cell types. Consult with the manufacturer if additional optimization is needed. - After nucleofection, add 80 µL of RPMI media to each small nucleocuvette well and let the cells sit at 37 °C for 15 min.
- Transfer the cell suspension into a well of a 24-well plate containing 400 µL of pre-warmed RPMI media (small nucleocuvette).
NOTE: Following nucleofection, cells are particularly fragile and should be transferred from the nucleocuvette to culture wells gently. - After day 2 post-nucleofection: follow surface marker staining protocol with CD55 antibody as described in section 3 and measure fluorescence of CD55 APC on a flow cytometer to assay epigenome editing efficacy.
NOTE: To assay nucleofection success, we recommend performing a nucleofection with GFP or mCherry mRNA. Two days post-nucleofection, perform flow cytometry of these cells to confirm successful nucleofection. We generally detect 90% to 100% of these cells expressing GFP or mCherry. An alternative is to perform a nucleofection with CRISPRi mRNA, which is known to robustly silence many target genes. When performing flow cytometry on day 2, CRISPRi treated samples should show target gene silencing in over 90% of cells.
3. Surface marker staining
NOTE: This section details quantifying the levels of CD55 protein after epigenome editing in K-562 cells. We quantify the decrease of CD55 expression in single cells using antibody staining and flow cytometry (see section 4 below) to evaluate CRISPRoff-mediated knockdown efficiency. Additional techniques, including reverse transcription quantitative PCR or western blotting, may also be utilized to confirm the level of knockdown at both transcript and protein levels.
- Count cells using an automated cell counter and add about 5.0 x 105 to 1.0 x 106 cells into a microcentrifuge tube and centrifuge cells at 500 x g for 5 min.
NOTE: More or fewer cells can be used for staining, however the concentration of antibodies may need to be scaled accordingly. - Discard the supernatant while ensuring not to disturb the pellet and add 500 µL of PBS and resuspend the pellet gently.
- Centrifuge the cells again at 500 x g for 5 min. Discard the supernatant and add the antibody at the desired concentration diluted in PBS. For CD55 staining, use 1 µL of Purified anti-human CD55 (0.5 mg/mL stock) in 49µL of PBS.
- Gently pipette the cells and allow them to incubate for 25 min in a dark location at room temperature. After incubation, spin the cells again at 500 x g for 5 min and discard the supernatant.
- Resuspend the pellet with 500 µL of PBS to wash off excess antibody. Centrifuge the cells at 500 x g for 5 min and discard the PBS supernatant. Resuspend in 100 µL to 200 µL of fresh PBS and add to a 96-well plate for flow cytometry analysis.
4. Flow cytometry
NOTE: This protocol is written for using a BD FACSymphony A1 Cell Analyzer. Specifics may vary depending on the flow cytometer that you are using. We suggest referencing the user manual for the machine that you are using for specifics.
- Before turning on the cytometry, check that the waste container is not full and that there is plenty of sheath fluid. Turn on the main power switch (green) that is near the sheath fluid.
- Turn on the plate reader using the back switch. Then, turn on the machine using the green button on the right side and make sure the machine is set to the 96-well plate reader function.
- Turn on the computer and log in. Open the software and log in using your username and password.
- Set up the experiment. Create a new experiment by selecting Experiment > New Experiment. Rename the experiment by double-clicking on the current name.
- Add a plate by clicking Experiment > New Plate. Select the well format to be used. To add specific wells to run, click on the well's location in the plate and then click the Blue Syringe (the well should now be highlighted in blue). Select multiple wells at the same time to add, if needed.
- Add specific names to each well by right-clicking on Specimen on the right of the plate display. Then select Experimental layout. This will open a page listing all the wells you selected. To rename the wells, click once and type in the desired name. Click OK when done.
- Add plots and appropriate gates to the experiment on the Global Worksheet. We recommend first plotting forward scatter area (FSC-A) vs side scatter area (SSC-A). Then create an arbitrary gate for live cells to later refine by clicking on the Polygon Tool.
- Right-click on this gate and click Drill Down. This will create a new plot only displaying cells from that live cell gate.
- Change the axis of this new plot to FSC-A vs forward scatter height (FSC-H) and create a gate for single cells.
- If constitutively expressing the sgRNA in cells, and the lentivirus also encodes for a fluorescent protein, then drill down this single cell gate and perform further gating for expression of that protein as a proxy for sgRNA expression (example: PE-CF594-A vs FSC-A).
- On the final setup gate (either for single cells or for sgRNA expressing cells), right-click and select Drill Down. Create a gate for assaying the expression of the reporter gene (example: BB515-A vs FSC-A).
- If using transfected cells and have a method for assaying transfection efficiency such as BFP expression, right click on the Reporter Expression Plot and click Duplicate. Change the axes to BV421-A vs FSC-A. Create an arbitrary gate for BFP-positive cells.
- Remove any parameters that are not in use in the Parameters tab of the cytometer display.
- To set up the machine and prepare it to run, click Run on the flow cytometer control panel. The button should appear green. In the software, go to HTS in the top panel and then Prime. Once finished, add the plate, making sure it is well positioned in the machine and the lid is OFF.
- To run, select all the well(s) to run and make sure controls are appropriately set. Select well by double clicking, change the sample volume to 10 µL and click Acquire Data in the Acquisition Dashboard. This will run the sample but will not record the data. As this sample is running, adjust the laser voltages as needed. Additionally, change the gates.
NOTE: We recommend running a sample that is negative for all colors being measured and one that is positive for all colors to make sure the laser parameters and gating are good. - After checking the laser settings, click on the Specimens in the Plate Panel and verify that the running values are set as desired.
- Click Run plate to run in the Acquisition Dashboard and record data from all wells. Once finished, clean the machine by removing the plate.
- To a new 96-well plate, add 10% bleach to the top four rows and H2O to the four wells below. Go to HTS > Clean. Once finished, close the program and turn off the buttons in the reverse order that they were turned on green side button > plate reader > green switch by waste.
- Add 20% bleach to the 96-well plate with cells and discard.
5. Data analysis
NOTE: This method outlines gating strategies and data processing to quantify epigenome editing by flow cytometry. The gating strategy is represented visually alongside example plots generated from the data analysis in Figure 2 and Figure 3.
- FlowJo gating setup
- Load all the FSC files onto FlowJo by dragging and dropping them into a new worksheet.
- Click on All Samples to begin making gates. This will ensure that the gates can easily be applied to all samples.
- Open a control sample (such as untransfected or un-nucleofected) by double-clicking on that file.
- Create a gate for Live Cells in a FSC-A vs SSC-A plot using the polygon tool. On the list of all the samples, a Live Cells tab should now appear below the sample name. Right-click on this gate and select Copy Analysis to Group to apply this gate to all samples (Figure 2A, Figure 3A).
- Double-click on the Live cells gate to drill down on the live cells. Change the axes to FSC-A vs FSC-H (Figure 2B, Figure 3B).
- Draw a polygon to gate for only single cells. Apply this gate to all samples as done in section 4 (Figure 2B, Figure 3B).
NOTE: If constitutively expressing a sgRNA construct with a fluorescent reporter, perform further gating on this color so that the analysis only focuses on cells that are expressing the sgRNA. An example of this would be creating a gate for mCherry positive cells (PE-CF594-A vs FSC-A) if the cells contain a sgRNA from the pLG1 backbone (Figure 2C, Figure 3C). - Double click on the Last Setup Gate (single cells or guide-expressing cells).
- If performing a plasmid transfection and the epigenome editor encodes for a BFP fusion, then create a gate for BFP positive cells (example: BV421-A vs FSC-A) for the Day 2 samples (Figure 2D).
- Create a gate for reporter expression (example: BB515-A vs FSC-A to gate for GFP negative cells) and apply this gate to all samples (Figure 2E, Figure 3D).
- Transfection analysis
- After performing the gating setup, click on Table Editor in the upper panel. Drag populations for analysis (examples: BFP positive, GFP negative).
- In the table editor panel, click Create Table. Copy these data into a spreadsheet to perform normalization calculations.
- Data processing
- Subtract the number of reporter negative cells (example: GFP negative) in the control sample from all other samples. There may be some background silencing of the reporter, so this removes this background noise from the data.
- All values will be normalized to the percentage of BFP positive cells (successfully transfected cells) measured on day 2. To do this normalization, take the value, divide by the percentage of BFP positive cells on day 2, and multiply by 100. This calculation yields the transfection efficiency normalized value of the edited cells. The data are now ready to be plotted. We recommend line graphs across an epigenome editing time course as depicted in Figure 2F.
- Nucleofection analysis
- After performing the gating setup, click on Table Editor in the upper panel. Drag populations for analysis (examples: APC negative).
- In the table editor panel, click Create Table. Copy these data into a spreadsheet editor to perform normalization calculations.
- Data processing
- Subtract the number of reporter negative cells (example: APC negative) in the control sample from all other samples. There may be some background silencing of the reporter, so this removes this background noise from the data. The data are now ready to be plotted. We recommend line graphs across an epigenome editing time course.
NOTE: Statistics may be used to quantify differences in silencing between two different epigenome editors. We recommend performing all transfection and nucleofection experiments in technical triplicates to perform statistical analyses. When depicting line graphs of silencing over time, plot standard deviation of technical replicates. Student t-tests at specific timepoints could be performed to compare silencing between two epigenome editors. Samples should be treated as unpaired with unequal variance.
- Subtract the number of reporter negative cells (example: APC negative) in the control sample from all other samples. There may be some background silencing of the reporter, so this removes this background noise from the data. The data are now ready to be plotted. We recommend line graphs across an epigenome editing time course.
Wyniki
For all epigenome editing experiments, proper controls are critical to assess epigenome editing efficiencies. We recommend using a control sgRNA, which does not target any sequence in the human genome. Using a non-targeting guide control will give confidence that changes at the target loci are driven by the epigenome editor being directed to that site rather than just from epigenome editor overexpression or non-specific binding. Additionally, for reporter gene-based experiments, we suggest using a dCas9-only control to ensure that changes in reporter expression are due to epigenome editor fusions rather than steric hindrance of the dCas9 binding to the target locus and temporarily impeding transcription (Figure 2F).
For transfection experiments, we recommend using an epigenome editor with an additional fluorescent protein fusion, such as BFP. This fusion allows for the visualization of cells successfully transfected with the epigenome editor through microscopy and flow cytometry. Successfully transfected cells will express high levels of BFP two days post-transfection (Figure 2D). The quantification of successfully transduced cells is used to normalize epigenome editing efficacy in later days (Figure 2F).
Both CRISPRoff and CRISPRi show peak silencing at day 5 post-transfection (Figure 2E-F). Different epigenome editors have distinct timelines of epigenome editing, such as heritable silencing with CRISPRoff and transient silencing with CRISPRi (Figure 2F). Figure 2F also displays the usage of dCas9 only as an important control for epigenome editing experiments. In mRNA nucleofection experiments, cells successfully edited with CRISPRoff will show strong silencing of the target gene by day 3 post-nucleofection (Figure 3E-F).
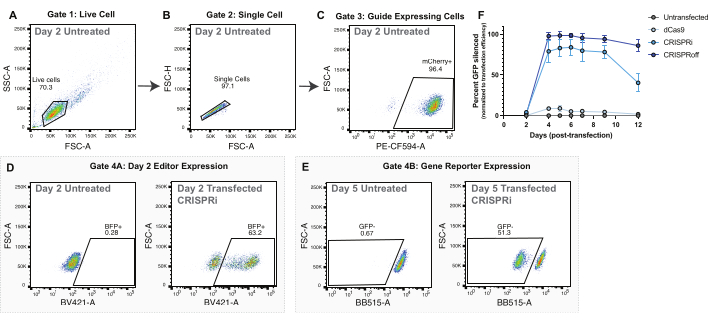
Figure 2: Gating strategy and representative data for epigenome editor delivery by plasmid transfection. (A-C) Representative flow plots to display gating strategy for plasmid transfection experiments. Displayed flow plots are of untransfected cells 2 days post-transfection. Each point in the plots represents one cell. (A) Flow plot of forward scatter area (FSC-A) and side scatter area (SSC-A) with a gate for live cells. (B) Flow plot of Live Cells displaying FSC-A and forward scatter height (FSC-H) with a gate for single cells. (C) Flow plot of Single Cells graphing PE-CF594-A (mCherry expression) and FSC-A. Gating for mCherry positive cells as a proxy for sgRNA expression. (D) Representative gating strategy for epigenome editor expression (BFP+) on day 2 post-transfection. The parent population is guide-expressing cells gated in (C). (E) Representative gating strategy for assaying silencing of CLTA-GFP reporter gene. Parent population is guide-expressing cells gated in panel C. (F) Silencing of CLTA-GFP over days post-transfection following plasmid delivery of dCas9, CRISPRi, and CRISPRoff. Percent CLTA-GFP silenced is normalized to transfection efficiency measured as BFP positive cells 2 days post-transfection. Points are averages of four transfection replicates. The error bars represent the standard deviation. Please click here to view a larger version of this figure.
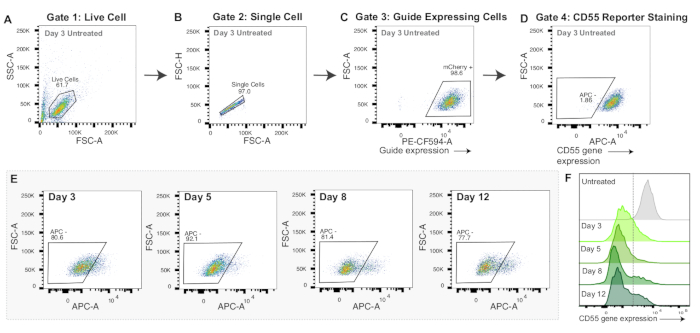
Figure 3: Gating strategy and representative data for CRISPRoff mRNA nucleofection. (A-D) Representative flow plots to display gating strategy for mRNA nucleofection experiments. Displayed flow plots are of control cells 3 days post-nucleofection stained with the APC anti-human CD55 antibody. Each point in the plots represents one cell. (A) Flow plot of forward scatter area intensity (FSC-A) and side scatter area intensity (SSC-A) with a gate for live cells. (B) Flow plot of Live Cells displaying FSC-A and forward scatter height intensity (FSC-H) with a gate for single cells. (C) Flow plot of Single Cells graphing PE-CF594-A (mCherry expression) and FSC-A. Gating for mCherry positive cells as a proxy for sgRNA expression. (D) Flow plot of APC-A versus FSC-A. Gate drawn for APC negative cells, which would indicate CD55 reporter gene silencing. (E) Flow plots of CD55 silencing (APC - gate) over 3, 5, 8, and 12 days post-nucleofection with CRISPRoff mRNA. (F) Overlaid histograms of CD55 protein expression (APC-A) over 3, 5, 8, and 12 days post-nucleofection with CRISPRoff mRNA compared to a stained but un-nucleofected control. Please click here to view a larger version of this figure.
| Plate | Seeding Density (cells per well) | Plasmid Amount |
| 96-well | 15,000 | 150 ng |
| 24-well | 90,000 | 500 ng |
| 6-well | 400,000 | 2 µg |
Table 1: Transfection scaling amounts. Seeding densities and plasmid DNA amounts for epigenome editor transfection into HEK293T cells at different scales.
Supplementary Table 1: sgRNA spacer sequences for epigenome editing experiments. sgRNA sequences for targeting epigenome editors to CLTA and CD55 along with a sequence for a non-targeting control guide. Additionally, oligos for cloning into the pLG1 backbone are listed. Please click here to download this File.
Supplemental File 1. Please click here to download this File.
Dyskusje
This protocol details two different transient delivery methods for CRISPR epigenome editors: plasmid DNA transfection and mRNA nucleofection. Both techniques have unique advantages, disadvantages, and general considerations (Figure 1F).
Plasmid DNA transfection leads to robust epigenome editor expression, and we include a BFP fusion within the epigenome editor constructs that allows for the detection and quantification of transfection efficacy using flow cytometry. Additionally, BFP expression can be used to sort for cells that express the epigenome editor or can be quantified to normalize silencing data at later time points, as detailed in this protocol. However, it is important to note that transfection efficiency is usually not 100%, and thus, the population will be heterogeneous for those that received the editor and those that did not, unless the cells are sorted. Furthermore, plasmid DNA delivery can trigger an immune response from cytoplasmic double-stranded DNA, leading to immune pathway activation and cytotoxicity. Finally, plasmid DNA transfection is not amenable to all cell types, and nucleofection of the plasmid may be necessary.
Unlike plasmid DNA transfection, mRNA nucleofection is applicable for many cell types and often results in high delivery efficacy. The editor mRNAs can be synthesized using commercially available in vitro mRNA synthesis kits one of which has been detailed previously51. Alternatively, mRNAs can be synthesized from Aldevron or Trilink. However, one caveat of mRNA nucleofection is that it does not produce enough editor protein to assay which cells received the epigenome editors. The experiments above suggest that almost every cell received the mRNA as we detect ~90% of target gene silencing at day 5 (Figure 3F). However, we recommend optimizing the pulse codes and mRNA-to-cell ratio for each cell type.
Another consideration is that epigenome editing initiated by plasmid DNA transfection or mRNA nucleofection has different silencing timelines. With plasmid DNA transfection, peak silencing with CRISPRoff and CRISPRi is observed on day 5 post-transfection (Figure 2F). In comparison, maximum silencing with CRISPRoff and CRISPRi delivered by mRNA nucleofection is seen as early as day 3 post-nucleofection (Figure 3F). Additionally, plasmid DNA persists in cells compared to mRNA, resulting in a longer duration of editor expression. Depending on experimental goals, a longer or shorter silencing timeline and expression duration may be preferred.
It is also important to note that epigenome editing cannot be universally applied to all genes or cell types. Inherent differences in genome sequence or chromatin states of the target genes may impact editor efficacy, even for the same gene in different cell types. For example, genes that lack annotated CpG islands can be difficult to stably silence by CRISPRoff-mediated DNA methylation48. Therefore, it may be necessary to test different epigenome editors to silence such genes. Additionally, we also recommend testing at least the top three CRISPRi gRNAs to identify the best guide for higher epigenome editing efficiency42. Nonetheless, given our limited toolbox of robust epigenome editors, it may be more effective to conduct gene knockout or use other knockdown strategies depending on the gene of interest.
This protocol is focused on CRISPRi and CRISPRoff, two of the many available CRISPR-based epigenome editors. Recent large-scale discovery studies have developed new tools for rewriting the human epigenome29,30,52. Epigenome editors have applications in biomedical research and therapeutics. For example, recent studies used DNA methylation and H3K9me3-based epigenome editing in mouse models and non-human primates, resulting in heritable repression of disease-associated gene53,54,55. We envision that future delivery modalities for epigenome editors will open new avenues for the broad application of epigenome editing.
Ujawnienia
J.K.N. is an inventor of patents related to the CRISPRoff/on technologies, filed by The Regents of the University of California.
Podziękowania
We thank the members of the Nuñez lab, especially Rithu Pattali and Izaiah Ornelas, for developing and optimizing the protocols described in this manuscript.
Materiały
| Name | Company | Catalog Number | Comments |
| 4D-Nucleofector | Lonza | AAF-1003 | |
| 96-well tissue culture plates | Corning | 3596 | |
| 96-well U-bottom Plate | Corning | 351177 | |
| APC anti-human CD55 Antibody | BioLegend | 311312 | |
| BD FACSymphony A1 Flow Cytometer | BD Biosciences | ||
| Bleach | Waxie | 11003428432 | |
| Centrifuge | Eppendorf | 5425 | |
| Countess Automated Cell Counter | Thermo Scientific | Countess 3 | |
| CRISPRoff transfection plasmid | Addgene | 167981 | |
| Diluent 2 Hematology Reagent for Flow Cytometry (Sheath fluid) | Thermo Scientific | 23-029-361 | |
| DMEM, High Glucose | Thermo Scientific | 11965118 | |
| DPBS | Gibco | 14-190-250 | |
| Eppendorf tubes | Thomas scientific | 1159M35 | |
| FBS | Avantor Seradigm | 89510-186 | |
| Lonza Walkersville SF Cell Line 4D-Nucleofector X Kit L | Fisher Scientific | NC0281111 | |
| mMESSAGE mMACHINE™ T7 ULTRA Transcription Kit | Thermo Fisher | AM1345 | |
| Opti-MEM | Gibco | 31985070 | |
| PCR strip tubes | USA Scientific | 1402-4700 | |
| Penicillin-Streptomycin-Glutamine | Gibco | 10378016 | |
| pLG1 sgRNA expression plasmid | Addgene | 217306 | |
| RPMI 1640 | Gibco | 22-400-105 | |
| SF Cell Line 96-well Nucleofector® Kit | Lonza | V4SC-2096 | |
| Tissue culture incubator | PHCbi | MCO-170AICUVDL-PA | |
| TransIT-LTI transfection reagent | Mirus | MIR 2306 | |
| Trypsin-EDTA (0.25%) | Gibco | 25200114 |
Odniesienia
- Kimura, H. Histone modifications for human epigenome analysis. Journal of Human Genetics. 58 (7), 439-445 (2013).
- Cong, L., et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 339 (6121), 819-823 (2013).
- Esvelt, K. M., Mali, P., Braff, J. L., Moosburner, M., Yaung, S. J., Church, G. M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature Methods. 10 (11), 1116-1121 (2013).
- Hou, Z., et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proceedings of the National Academy of Sciences. 110 (39), 15644-15649 (2013).
- Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533 (7603), 420-424 (2016).
- Gaudelli, N. M., et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 551 (7681), 464-471 (2017).
- Anzalone, A. V., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 576 (7785), 149-157 (2019).
- Anzalone, A. V., et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nature Biotechnology. 40 (5), 731-740 (2022).
- Zhang, X. -H., Tee, L. Y., Wang, X. -G., Huang, Q. -S., Yang, S. -H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy - Nucleic Acids. 4, e264(2015).
- Lin, Y., et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Research. 42 (11), 7473-7485 (2014).
- Adikusuma, F., et al. Large deletions induced by Cas9 cleavage. Nature. 560 (7717), E8-E9 (2018).
- Kosicki, M., Tomberg, K., Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nature Biotechnology. 36 (8), 765-771 (2018).
- Cullot, G., et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nature Communications. 10 (1), 1136(2019).
- Fiumara, M., et al. Genotoxic effects of base and prime editing in human hematopoietic stem cells. Nature Biotechnology. 42 (6), 877-891 (2024).
- Tsuchida, C. A., et al. Mitigation of chromosome loss in clinical CRISPR-Cas9-engineered T cells. Cell. 186 (21), 4567-4582.e20 (2023).
- Nakamura, M., Gao, Y., Dominguez, A. A., Qi, L. S. CRISPR technologies for precise epigenome editing. Nature Cell Biology. 23 (1), 11-22 (2021).
- Villiger, L., Joung, J., Koblan, L., Weissman, J., Abudayyeh, O. O., Gootenberg, J. S. CRISPR technologies for genome, epigenome and transcriptome editing. Nature Reviews Molecular Cell Biology. 25 (6), 464-487 (2024).
- McCutcheon, S. R., Rohm, D., Iglesias, N., Gersbach, C. A. Epigenome editing technologies for discovery and medicine. Nature Biotechnology. 42 (8), 1199-1217 (2024).
- Perez-Pinera, P., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature Methods. 10 (10), 973-976 (2013).
- Maeder, M. L., Linder, S. J., Cascio, V. M., Fu, Y., Ho, Q. H., Joung, J. K. CRISPR RNA-guided activation of endogenous human genes. Nature Methods. 10 (10), 977-979 (2013).
- Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S., Vale, R. D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 159 (3), 635-646 (2014).
- Gilbert, L. A., et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 154 (2), 442-451 (2013).
- Alerasool, N., Segal, D., Lee, H., Taipale, M. An efficient KRAB domain for CRISPRi applications in human cells. Nature Methods. 17 (11), 1093-1096 (2020).
- Nakamura, M., Gao, Y., Dominguez, A. A., Qi, L. S. CRISPR technologies for precise epigenome editing. Nature Cell Biology. 23 (1), 11-22 (2021).
- Tycko, J., et al. High-Throughput Discovery and Characterization of Human Transcriptional Effectors. Cell. 183 (7), 2020-2035.e16 (2020).
- Alerasool, N., Leng, H., Lin, Z. -Y., Gingras, A. -C., Taipale, M. Identification and functional characterization of transcriptional activators in human cells. Molecular Cell. 82 (3), 677-695.e7 (2022).
- Ludwig, C. H., et al. High-throughput discovery and characterization of viral transcriptional effectors in human cells. Cell Systems. 14 (6), 482-500.e8 (2023).
- DelRosso, N., et al. Large-scale mapping and mutagenesis of human transcriptional effector domains. Nature. 616 (7956), 365-372 (2023).
- Tycko, J., et al. Development of compact transcriptional effectors using high-throughput measurements in diverse contexts. Nature Biotechnology. , (2024).
- Wilson, C. M., et al. Combinatorial effector targeting (COMET) for transcriptional modulation and locus-specific biochemistry. , (2024).
- Liu, X. S., et al. Editing DNA Methylation in the Mammalian Genome. Cell. 167 (1), 233-247.e17 (2016).
- Bintu, L., et al. Dynamics of epigenetic regulation at the single-cell level. Science. 351 (6274), 720-724 (2016).
- Hilton, I. B., et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology. 33 (5), 510-517 (2015).
- Cano-Rodriguez, D., et al. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nature Communications. 7 (1), 12284(2016).
- O'Geen, H., et al. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Research. 45 (17), 9901-9916 (2017).
- O'Geen, H., et al. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics & Chromatin. 12 (1), 26(2019).
- Kim, J. -M., et al. Cooperation between SMYD3 and PC4 drives a distinct transcriptional program in cancer cells. Nucleic Acids Research. 43 (18), 8868-8883 (2015).
- Kearns, N. A., et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nature Methods. 12 (5), 401-403 (2015).
- Li, K., et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nature Communications. 11 (1), 485(2020).
- Stepper, P., et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Research. 45 (4), 1703-1713 (2017).
- Valbuena, R., et al. Prediction and design of transcriptional repressor domains with large-scale mutational scans and deep learning. , (2024).
- Horlbeck, M. A., et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760(2016).
- Liu, Y., et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell. 23 (5), 758-771.e8 (2018).
- Black, J. B., et al. Master Regulators and Cofactors of Human Neuronal Cell Fate Specification Identified by CRISPR Gene Activation Screens. Cell Reports. 33 (9), 108460(2020).
- Yang, J., et al. Genome-Scale CRISPRa Screen Identifies Novel Factors for Cellular Reprogramming. Stem Cell Reports. 12 (4), 757-771 (2019).
- Chakraborty, S., Ji, H., Kabadi, A. M., Gersbach, C. A., Christoforou, N., Leong, K. W. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem Cell Reports. 3 (6), 940-947 (2014).
- Black, J. B., et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell. 19 (3), 406-414 (2016).
- Nuñez, J. K., et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 184 (9), 2503-2519.e17 (2021).
- Xu, X., et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery. 2 (1), 16009(2016).
- Leonetti, M. D., Sekine, S., Kamiyama, D., Weissman, J. S., Huang, B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proceedings of the National Academy of Sciences. 113 (25), (2016).
- Pattali, R. K., Ornelas, I. J., Nguyen, C. D., Xu, D., Divekar, N. S., Nuñez, J. K. CRISPRoff epigenetic editing for programmable gene silencing in human cells without DNA breaks. , (2024).
- Moon, H. C., Herschl, M. H., Pawluk, A., Konermann, S., Hsu, P. D. A combinatorial domain screening platform reveals epigenetic effector interactions for transcriptional perturbation. , (2024).
- Cappelluti, M. A., et al. Durable and efficient gene silencing in vivo by hit-and-run epigenome editing. Nature. 627 (8003), 416-423 (2024).
- Naumann, E. N., et al. rainwide silencing of prion protein by AAV-mediated delivery of an engineered compact epigenetic editor. Science. 384 (6703), ado7082(2024).
- Tremblay, F., et al. A potent epigenetic editor targeting human PCSK9 for durable reduction of low-density lipoprotein cholesterol levels. Nature Medicine. 31 (4), 1329-1338 (2025).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone