Method Article
Comprehensive Analysis of Drug Response using the FLICK Assay
In This Article
Summary
This protocol describes how to use the FLICK assay for evaluating drug responses, including detailed instructions for using this assay to compute the drug-induced growth rates and death rates and to evaluate the mechanism of drug-induced cell death.
Abstract
For understanding drug efficacy, a critical need is to characterize the extent of drug-induced cell death. Efforts to quantify the level of drug-induced cell death are challenged by the existence of more than a dozen molecularly distinct forms of regulated death, each with its own activation timing and biochemical hallmark features. Furthermore, for some necrotic death subtypes, hallmark features are only observed transiently and are rapidly lost due to cell rupture. Thus, even when using a combination of death pathway-specific assays, it is challenging to accurately quantify the total amount of cell death or the relative contributions of each death subtype. Another issue is that many death-specific assays ignore how drugs affect cell proliferation, making it challenging to interpret if a drug-treated population is expanding or shrinking. The FLICK assay allows for quantification of the total level of cell death following stimulation in a manner that is specific to death but also largely agnostic to the type(s) of death activated. Additionally, the FLICK assay retains information about the total population size and cell proliferation rate. In this manuscript, we describe the basic use of the FLICK assay, how to troubleshoot this assay when using different types of biological material, and how to use the FLICK assay to quantify the contributions of each type of cell death to an observed drug response.
Introduction
For anti-cancer drugs, pre-clinical evaluation of drug sensitivity generally involves testing how drugs affect the viability of cells in culture1. Cell viability following drug exposure is a product of at least two separate effects: drug-induced inhibition of cell proliferation and activation of cell death2. Unfortunately, although cell death is a critical feature that is required for durable drug responses, standard approaches fail to clarify the degree to which a drug activates cell death3.
Common drug response assays include those that directly count cells (e.g., Coulter Counter, some uses of flow cytometry or microscopy), quantify the ability of cells to proliferate (e.g., colony formation assay), or quantify a metabolic activity (e.g., CellTiter-Glo, tetrazolium based MTT or MTS assays). A shared feature of these assays is that the data generated is proportional to the number of live cells. Because drugs vary considerably in how they coordinate growth inhibition and cell death, the number of live cells following drug exposure provides an unreliable insight into the level of drug-induced cell death3. Furthermore, because cancer cells generally proliferate rapidly in cell culture, the number of live cells can be dramatically reduced relative to the untreated population without inducing any cell death4. Thus, a central flaw is that the degree of cell death cannot be quantified without measuring both the number of live and dead cells.
Quantifying the number of dead cells following drug exposure is challenging to do accurately for several reasons2. First, more than a dozen regulated cell death pathways exist5,6,7,8. Although biochemical markers generally exist for identifying each type of regulated cell death, these markers vary in their specificity, and no single assay can be used to simultaneously quantify all death subtypes. Secondly, the timing of activation for each form of cell death can vary quite dramatically depending on the context, so a complete picture cannot emerge unless death is quantified over time9,10. Many biochemical assays for quantifying cell death produce an end-point measurement, so generating kinetic data can be challenging and limited by cost. A third complication is that the dead cell itself is a transient intermediate state between the live cell state and dissociated cell debris. The stability of dead cells varies depending on the death subtype, with some types, such as apoptosis, creating relatively stable corpses, whereas other types of death cause rapid lysis. Thus, methods of quantifying death that require collection and counting of dead cells also will produce a biased understanding of cell death. Finally, a fourth limitation is that biochemical assays that quantify the degree of cell death typically fail to provide any insight into how a drug alters proliferation. Thus, the overall population size - and, importantly, whether the population is expanding or shrinking - cannot be interpreted.
Some microscopy-based assays, such as STACK and SPARKL, are effective at measuring live and dead cells over time, and these assays can produce comprehensive insights about drug-induced cell death10,11. These assays, however, require specialized instruments, such as the Incucyte microscope, creating limitations in throughput and access to these approaches. Additionally, microscopy-based techniques require that dead cells remain in the focal plane of the microscope throughout the duration of the experiment, compromising the ability to quantify dead cells when they lose adherence from the plate or over time as dead cells decay. Similarly, microscopy-based assays face challenges when applied in the context of suspension cultures, as cells drift in and out of a given focal plane.
To address the issues highlighted above, we have generated an assay called FLICK (Fluorescence-based and Lysis-dependent Inference of Cell Death Kinetics)12,13. The goal of the FLICK assay is to determine the level of drug-induced cell death, regardless of how the cells are dying. The FLICK method uses cell impermeant dyes, whose fluorescence depends on DNA binding. A key feature of FLICK is the use of these fluorophores to label dead cell accumulation over time by virtue of their accessible DNA, followed by a mechanical detergent-based lysis to permeabilize any live cells at the end of the assay. These data, combined with mathematical modeling, enable the quantification of both live and dead cell populations with continuous temporal resolution and without requiring the collection or handling of dead cells. Furthermore, the use of a plate reader to evaluate dead cell fluorescence allows for the evaluation of dead cells without requiring that dead cells remain intact, thus alleviating a bias against necrotic forms of death that result in cell rupture. Finally, the FLICK assay requires minimal plate handling and can rapidly generate kinetic measurements, allowing for high-throughput drug screening. In this protocol, we focus on the use of the FLICK assay, including how to use FLICK to infer the drug-induced growth rate, death rate, and/or the mechanisms of cell death.
Protocol
1. Optimization of permeabilization time for each cell line of interest
NOTE: The volumes and amounts described are for optimizing one cell line. These values should be scaled up based on the number of cell lines that are to be tested.
- Plate the desired number of cells in each well of a 96-well, optical bottom, black-walled plate. Add 100 µL of complete medium and let the cells adhere to the plate overnight.
NOTE: The plated cell number should be optimized with consideration for the growth rate of the cells, optimal densities, and length of the assay. Typical starting cell numbers for adherent cancer cell lines measured over 72 h are 1500 - 5000 cells per well. For instance, U2OS cells may be plated at 2000 cells per well in DMEM with 10% FBS, 2 mM glutamine, and 1% Pen-Strep and cultured overnight in standard conditions (5% CO2, 37 °C, with humidity). - In a 15 mL conical centrifuge tube, prepare 1.5 mL of a 1.5% solution of Triton-X in phosphate buffered saline (PBS). Vortex the 1.5% Triton-X solution for 5 s at max speed. Place in a 37 °C water bath for at least 30 min.
- Visually inspect the solution to ensure that Triton-X has fully dissolved by vortexing for several seconds. Ensure that the solution is homogenous.
- Add 10 µL of the 1.5% Triton-X solution to each well of the plated cells. Do not mix.
NOTE: At this stage, it is not desirable to mix using repeated pipetting due to the formation of bubbles. - Return cells to the 5% CO2, 37 °C with humidity cell culture incubator for at least 1 h.
NOTE: Following application of the Triton-X solution, most cell lines are fully lysed in 2-3 h. Waiting up to 24 h has minimal impact on fluorescence signal when using dyes such as SYTOX Green. Increasing the percentage of Triton-X may shorten incubation time for difficult to lyse cell lines. - Observe cell morphologies on a light microscope using a 10x objective. Inspect cells once every hour until cell bodies are no longer visible. Record the time required for cell permeabilization.
NOTE: If a fluorescent microscope is available, lysis can be visualized using a cell-impermeable DNA dye, such as SYTOX Green. The dye can be added at a 10x final concentration to the Triton-X solution, and permeabilization can be confirmed by fluorescence microscopy.
2. Selection and calibration of DNA stain
NOTE: A requirement for the FLICK assay is the use of a cell impermeant fluorophore that emits a signal in a DNA-binding dependent manner, does not affect cell viability, and produces a signal that scales linearly with cell number. This protocol uses SYTOX Green. Other dyes with similar properties may also be suitable for the FLICK assay, but these should each be evaluated and calibrated. See Table 1 for examples.
- For the selected DNA dye, determine the range of concentrations to be tested based on the manufacturer's recommendation.
- For each concentration to be tested, plate 40,000 cells in 180 µL of cell culture medium in triplicate along the leftmost column of a 96-well, optical bottom, black-walled plate. Add 90 µL of medium to the remaining wells.
NOTE: This will be used to generate a linear cell titration across the rows of the plate. The final concentration of the first wells will be 20,000 cells/well. Optical bottom plates are not required for plate readers that read fluorescence from the top of the well. The cell culture medium should be the same medium used for growing the cells being tested. For instance, if using U2OS cells, 40,000 cells should be resuspended in 180 µL of DMEM. - Using a multichannel pipette, create a 1:2 serial dilution by transferring 90 µL from the leftmost column into the adjacent column to the right. Pipette 15x to mix.
- Repeat step 2.3, moving from the second column to the third column, then from the third column to the fourth column, and so on. Finish the titration in the second to last column of the plate. Leave the last column without cells so the background signal of cell culture media can be obtained.
- For the second-to-last column, remove 90 µL so all wells have 90 µL of medium containing a varied number of cells. Let cells adhere for 6 h in a 37 °C cell culture incubator.
- Prepare a 10x solution of DNA dye for each concentration of dye to be tested, with the DNA dye diluted into complete growth media used for culturing the cells of interest. Prepare 1.5 mL of a 1.5% Triton-X solution in PBS, as described in step 1.
- To each well of the 96-well plate, add 10 µL of the 10x DNA dye. Add 10 µL of 1.5% Triton-X solution to each well to permeabilize cells. Incubate cells in this solution for the optimal time, which was determined in step 1.
NOTE: After the addition of Triton-X, the final concentration of DNA dye is slightly less than 1x, but this is inconsequential. Following incubation, the entire plate should be lysed. The manual cell titration will be used to identify the optimal DNA dye concentration and the optimal settings for acquiring fluorescence. - Measure fluorescence intensities across the cell titration plate. Quantify fluorescence over a range of acquisition settings as outlined by the manufacturer of the DNA dye. Depending on the plate reader used, modify the excitation and emission wavelengths and/or the digital gain.
- Remove the background signal from each measurement by subtracting the average signal of the column containing no cells. These values can be found in the right-most column of wells in the serial dilution plate.
- Determine the linearity of each concentration of DNA dye for each acquisition setting. Linearity can be determined by graphing the cell number against the fluorescence signal and performing linear regression. The coefficient of determination (r2) reports the degree to which the fluorescence signal is linearly related to the cell number.
- Pick the acquisition settings and DNA dye concentration that has the best combination of linearity and dynamic range.
NOTE: Consider the entire range of cell numbers from 0 to 20,000 cells. Ensure that the gain and DNA dye concentration are linear at low cell numbers, between 0 and 1000 cells. Several instrument settings or DNA dye concentrations may provide a strong correlation over the whole range. However, it is essential that the low-end sensitivity is robust, such that small changes in dead cells can be accurately measured.
3. Cell Plating, In-Well Drug Application, and Dead Cell Fluorescence Measurement Over Time in Drug-Treated Plates
NOTE: Drug dilution plates can be designed flexibly based on experimental needs. Generally, drug dilution plates will include a log or semi-log dilution series of one or several drugs.
- Consider the optimal plating layout for the experiment. Avoid measurements from the outer wells of a 96-well plate to decrease noise associated with growth variation, as the outer wells experience different temperatures, oxygenation, and evaporation compared to the inner wells of the plate.
NOTE: Including controls throughout the landscape of the plate will increase the robustness of the growth data. Consider including control wells in the center and edges of the plate. - Determine the number of plates required for the experiment. Include one additional plate to be lysed at the beginning of the assay to establish the average cell number per well at the time of drugging (i.e., the T0 control plate).
- Prepare a cell suspension in complete growth media.
- Based on the number of plates needed for the experiment (calculated in step 3.2), determine the volume of the cell suspension required. If seeding plates at 90 µL per well, about 10 mL is required per plate. Multiply the number of plates required by 10 mL.
NOTE: It is desirable to increase this volume to account for the dead volume of the reservoir and volume loss during pipetting. For instance, the calculated volume may be multiplied by 1.2. - Count cells and prepare a cell dilution based on the desired number of cells per well. For instance, if aiming to seed 2000 cells in 90 µL of media per well of a 96-well plate, for 3 total plates, calculate the total number of cells needed for the experiment as below:
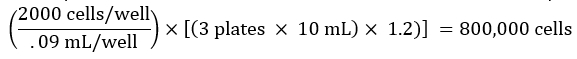
Similarly, if the counted cell suspension is 400,000 cells/mL, the volume of the cell suspension needed for the experiment would be:
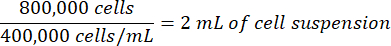
- Correct the volume of cell culture media used for plating by subtracting the volume of the counted cell suspension to be added.
36 mL media - 2 mL cell suspension = 34 mL corrected media volume
- Based on the number of plates needed for the experiment (calculated in step 3.2), determine the volume of the cell suspension required. If seeding plates at 90 µL per well, about 10 mL is required per plate. Multiply the number of plates required by 10 mL.
- Mix the counted cell suspension with the correct media volume using a serological pipette. Transfer this cell suspension to a V-bottom reagent reservoir.
- Using a multichannel pipette, add 90 µL of the cell suspension to each well of the 96-well plates. Mix the cell suspension in the reagent reservoir regularly using repeated pipetting with a serological pipette to ensure that the desired to ensure that the desired concentration of cells per volume is maintained.
- Let cells adhere overnight in a 37 °C cell culture incubator (generally 12-24 h).
- Prepare a 1.5% Triton-X solution in PBS, as described in step 1. The volume of this solution should be sufficient to lyse all plates. About 1.5 mL of 1.5% Triton-X in PBS is sufficient to lyse one 96-well plate.
NOTE: The 1.5% Triton-X solution in PBS is stable for over a week and can be made in advance for convenience. - Determine the amount of cell culture media required for making a drug dilution plate. Prepare drug dilution plates in U- or V-bottom 96-well clear plates.
- To ensure that the volume of diluted drug is sufficient for the experiment, increase the minimum required volume to account for volume loss during pipetting. For example, if drugs are to be added in 10 µL to each well of a 96-well plate and the experiment includes three plates, 30 µL of diluted drug are required per well. Prepare 40-50 µL of drugged media per well to account for volume loss. Add 1.5 mL to the volume of media required to account for the media needed for the T0 control plate.
- Prepare a 10x concentration of the selected DNA dye in complete growth media. This concentration is based on the concentration selected in step 2.11. The total volume of this solution is calculated in step 3.8.
- Using the DNA dye + growth medium from step 3.9, prepare a 10x concentration of each drug to be tested. Create only the highest dose of each drug in the drug dilution plate and serially dilute the drugs with a multichannel pipette, mixing 15x between each well.
NOTE: When making the highest dose for a given drug in the drug dilution plate, the volume of the drug should be accounted for to ensure that the concentration is accurate. - Using a multichannel pipette, add 10 µL of drug + DNA dye solution from step 3.10 to the plates containing cells.
NOTE: For drug dilution plates in which a serial dilution was created, if drugs are added to the cells starting with the lowest concentration of drug and working up to the highest concentration, the tips of the multichannel pipette do not need to be changed. However, tips on the multichannel pipette should be changed between each plate and whenever the tips were used in a well containing a different drug or a higher concentration of the drug. - Read the fluorescence for all plates in which drugs were added. This fluorescence reading is the T0 reading (i.e., dead cells at Time = 0 h).
- Return plates to the incubator after taking the T0 fluorescence reading.
- Add 10 µL of the 10x DNA dye solution created in step 3.9 and 10 µL of the 1.5% Triton-X solution in step 3.7 to the T0 control plate. Return this plate to the incubator for the amount of time selected in step 1.8.
- Read the T0 control plate once the cells are fully lysed, as described in step 1.6.
4. Measure the dead cell fluorescence over time for drug-treated plates
NOTE: Minimize the time plates are out of the incubator. Prolonged changes in temperature can affect cell viability, and exposure to light can compromise DNA fluorophores, such as SYTOX Green.
- Acquire fluorescence readings for all drug-treated plates every 3-4 h after drugging. Plates do not need to be read overnight unless very precise death kinetics are required.
NOTE: Time points do not need to be taken at fixed/standard intervals. In general, time points can be taken exclusively during normal working hours without causing errors in the downstream analysis of death kinetics. The kinetics can be inferred as long as some measurements are taken in the period before death onset, during the rising phase of death, and during the saturation/plateau phase. - At the desired final time point, acquire a fluorescence reading. Immediately afterward, add 10 µL of the 1.5% Triton-X solution prepared in step 3.7 to lyse the cells.
- Return the plates to the incubator and allow the cells to permeabilize for the time determined in step 1.
- Acquire fluorescence readings following permeabilization. This fluorescence value is proportional to the total number of cells (i.e., live + dead cells) for each well.
5. Calculate the lethal fraction kinetics
NOTE: Calculations described in this protocol can be analyzed in any format or software. However, using a programming environment such as MATLAB, R, or Python will allow for faster and more flexible analysis.
- Calculate the average fluorescence values from the T0 control plate using the 50% trimmed mean. This value is proportional to the total number of cells prior to drugging.
- Using curve fitting and an exponential growth function, calculate the kinetics of population growth for all wells in the experiment. The initial population size for each well is the average T0 fluorescence value calculated in step 5.1. The final population size for each well is the post-permeabilization value calculated in step 4.4. The duration of the assay is from 0 h to the assay endpoint selected in step 4.4.
- Using the growth parameters determined in step 5.2, calculate the number of total cells at every measured time point in the assay for every well. Determine the number of live cells at each measured time point by subtracting the dead cell measurement from the total cells calculated in step 5.2.
NOTE: Due to small amounts of noise in the assay, occasionally, the live cell number may be a small negative number. This can be manually set to 0 since there cannot be a negative number of live cells and most likely indicates all cells in that well were dead. - Calculate lethal fraction (LF) for each time point by dividing the dead cell fluorescence signal by the total cell signal for each time point.
- Fit a Lag Exponential Death (LED) equation to the lethal fraction time course data10. To avoid arbitrary kinetic parameters for doses of a drug that do not induce any significant lethality, fit to a linear model with a slope equal to 0. Determine significant levels of lethality based on the noise in the assay or the LF observed for undrugged conditions.
- Record death onset time (DO) from the LED equation. The LED equation has four parameters: the initial LF (LFi), the LF at plateau (LFp), the initial death rate (DR), and the DO. Infer these parameters from the data using a non-linear regression (i.e., curve fitting).
- Compute the fractional viability (FV) at the assay endpoint for each drug at each dose. Subtract the endpoint LF value from 1 or divide the number of live cells by the total cells.
NOTE: FV can be calculated at any time point, not only at the assay endpoint. These data are used to evaluate dose pharmacology (e.g., drug IC50 or EC50).
6. Calculate the GR value
- Determine the average number of live cells for each well at the start of the assay. Calculate this value using the post-permeabilization T0 reading calculated in step 3.15 and subtracting the T0 reading for each well, which was collected in step 3.12. In the GR equation below (step 6.3), this value is referred to as x0.
NOTE: Occasionally the highest dose of a drug can interfere with the fluorescence reading of the DNA dye. It may be desirable to compute the average number of live cells at time 0 using the control wells. - For the control wells (xctrl), determine the average number of live cells for each well at the assay endpoint. Determine the average number of live cells at the assay endpoint for each drug treated condition (x(c)).
NOTE: Depending on the variation in cell growth across the plate, different xctrl values can be used for normalization of each well. For instance, if the xctrl values vary systematically across the plate, it may be desirable to use the closest xctrl value rather than the plate's average xctrl value. - For each drug-treated well, compute the normalized growth rate inhibition value (GR) using the following equation4:
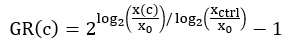
Perform curve fitting using a 4-parameter logistic regression. GR is on a scale from -1 to 1.
7. Calculate drug-induced growth and death rates using the GRADE method
NOTE: GR represents the net population growth rate, not the true cell proliferation rate. The drug-induced population growth and death rates can be computed using a combination of the GR and the lethal fraction (LF).
- Determine the doubling time (τ) of the cell line during the experiment using the average number of live cells from the T0 control reading (x0), the average number of live cells from the control condition at assay endpoint (xctrl), and the length of the assay in hours (t) to solve the following equation:

NOTE: Accurate calculation of cell doubling time (τ) requires that cells continuously double throughout the length of the assay. For cells that experience contact inhibition, cells should be seeded at starting densities that will not become confluent during the desired assay time. For example, U2OS has an average doubling time of 24 h. Seeding 2000 cells per well in a 72 h assay is optimal for interpreting their growth rate. - Determine the relationship between growth/death rates and population size. To do this use a birth/death model and a simulation initialized with all plausible pairwise combinations of growth and death rates. This simulation requires the average number of live cells from the T0 control plate (x0), the length of the assay (t), and a user- defined range of plausible proliferation rates (p), and death rates (d).

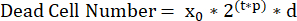
- To determine plausible values for growth rates, start with the untreated growth rate in cell population doublings per h (1/T) as the highest growth rate and 0 as the lowest growth rate. Divide this range into 500 equally spaced segments. A similar operation can be applied for the death rate, testing a range from 0 to 1.
NOTE: All pairwise combinations of 500 rates can be performed using any programming environment but may be too computationally intense if using some software. Reducing this to all pairwise combinations of 50 growth and death rates can alleviate this issue, with a reduction in the precision of the inference. A spreadsheet template has been provided with this protocol.
- To determine plausible values for growth rates, start with the untreated growth rate in cell population doublings per h (1/T) as the highest growth rate and 0 as the lowest growth rate. Divide this range into 500 equally spaced segments. A similar operation can be applied for the death rate, testing a range from 0 to 1.
- Determine the GR and LF values (described above) for each simulated proliferation (p) and death (d) rate pair.
NOTE: Steps 7.2-7.3 will generate a table that contains each theoretical pair of proliferation and death rates, with the computed GR and LF values for that theoretical pair. This will function as a look up table to associate an experimentally observed pair of GR and LF values with a pair of drug-induced proliferation and death rates. Testing more than 500 discrete values for proliferation and death rates will result in a more numerically precise value, but this level of precision will likely be beyond the precision of the assay. - Compute the pairwise distance between each experimentally calculated GR/LF pair and each theoretical GR/LF pair in the look up table. Infer the theoretical pair with the minimum distance to the experimentally observed pair of GR/LF values to be the true drug-induced proliferation and death rates.
8. Determination of death pathways using pathway-selective chemical inhibitors
NOTE: Chemical inhibitors alone are insufficient for definitively determining the mechanism of death for a given drug. Chemical inhibitors of death pathways should be used to determine which biochemical or phenotypic responses should be explored in subsequent experiments, which are likely to include morphological assessment, pathway-specific biochemical markers, and evaluation of genetic dependencies.
- Optimize the dose of each cell death inhibitor in the cell line of interest by testing a dose range of inhibitor concentrations in the context of a canonical activator of the death pathway(s) of interest (Table 2). Ideally, the selected dose of inhibitor should not affect cell viability on its own (check the GR metric).
NOTE: Not all cell lines can activate all death pathways. Biochemical or phenotypic validation may be required when testing activators/inhibitors of different death pathways. - Consider the optimal plating layout for an inhibitor screen. To minimize batch effects, keep each drug on the same plate as the inhibitor(s) that are being evaluated, with replicates on separate plates.
- Seed the desired number of cells in each well of a 96-well, optical-bottom, black-walled plate, similar to step 3.3. Decrease the plating volume to 80 µL to account for the volumes of the inhibitor and drug.
- Prepare 10x concertation of the inhibitors to be tested in complete media and add 10 µL to each well. Pre-treat cells with death pathway inhibitors for 2-4 h. Afterward, add the drugs of interest and acquire fluorescence reading, as in steps 3-5.
- Evaluate the change in death onset time (DO) and/or the max lethal fraction to assess the effectiveness of the inhibitor(s). The DO is an inferred parameter from the Lag Exponential Death (LED) model. See steps 5.6 - 5.7.
Results
Using this protocol, we explored the sensitivity of U2OS cells to the HDAC inhibitor Belinostat. These experiments were performed using 2 µM SYTOX Green to label dead cells (Figure 1A). Kinetic readings were made using a fluorescent plate reader at a 130 gain setting (Figure 1B). Cells were lysed in 1.5% Triton-X solution in PBS for 2 h at the end of the assay (Figure 1C-D).
The FLICK protocol produces insights into the population growth rate and drug-induced cell death rate. These insights can be viewed separately or using the drug GRADE visualization and analysis framework, viewed together (Figure 1E). Evaluating Belinostat sensitivity using the GR value reveals that 1 µM Belinostat results in a GR value of approximately 0 (Figure 2A). On the GR scale, positive values report the rate of population expansion, and negative values report the rate of population shrinkage. Thus, a GR value of 0 reveals that the population remains at stasis (i.e., a cytostatic drug response). Given the conventional interpretation of cytostasis, one may conclude that 1 µM Belinostat causes full growth inhibition without any cell killing. However, evaluating the drug-induced lethal fraction reveals that 1 µM Belinostat caused roughly 50% lethality over the course of this assay (Figure 2B). In most cases, these two insights would be derived from different experiments that cannot be compared in an apples-to-apples manner. Importantly, using the FLICK assay, both insights are derived from the same experimental data but analyzed differently to capture the effects of Belinostat on population growth versus cell death.
These two insights can also be integrated using the GRADE analytical method (Figure 2C). Drug GRADE juxtaposes the Lethal Fraction, which is proportional to the average death rate, against the GR value, the net population growth rate. Integrating the data in this way provides a visualization of how a drug coordinates growth inhibition and death activation. Within the GRADE plot: drugs that inhibit growth without activating death (i.e., cytostatic drugs, using the common interpretation) will occupy the top boundary, drugs that only kill without altering cell proliferation rates will occupy the right boundary, and drugs that first cause growth inhibition, followed by the death of growth-arrested cells will occupy the left boundary (i.e., bi-phasic drugs; Figure 2C). Drugs that result in GR/LF responses that fall within these boundaries can be inferred to simultaneously inhibit proliferation to some extent while also activating cell death (i.e., coincident drugs)3. Importantly, the position of each drug response within the GR/LF space can be used to compute the true proliferation rate (p) and average death rate (d) for each concentration of a drug (Figure 2D-E). Note that the GR value is not the true cell proliferation rate but rather the net population growth rate (i.e., the net effect combining the true proliferation rate and the drug-induced death rate). Also, the death rate produced in a GRADE analysis (d, which is the average death rate) is distinct from the death rate parameter in the LED kinetic analysis (DR, the maximum velocity or maximum death rate).
The FLICK assay can generate death-specific data, such as the drug-induced lethal fraction; however, these data are agnostic to the mechanism of cell death. To gain insights into the mechanism of cell death using FLICK, the simplest method is to determine how lethal fraction kinetics are altered by the inclusion of death pathway-specific inhibitors. Inhibiting a relevant death mechanism should result in decreasing the drug-induced lethal fraction and/or delaying the death onset time. Here, we show that 50 µM of the pan-caspase inhibitor z-VAD rescues about 50% of the lethality induced by 1 µM Belinostat (Figure 2F). Importantly, like drugs, inhibitors have limited specificity and may accidentally inhibit or exacerbate other death mechanisms. Thus, before a definitive conclusion is made, these data should be complemented with other insights, such as cell morphology, measurement of biochemical markers of activation, and/or evaluation of pathway-specific genetic dependencies14.
To formally illustrate how the interpretation of a drug response may be altered by a FLICK-based evaluation and GRADE-based analysis, we next explored three drugs that feature different types of growth/death coordination. Palbociclib is a Cdk4/6 inhibitor that results in a cytostatic response (growth inhibition with no cell death). Camptothecin is a topoisomerase I inhibitor that causes a bi-phasic response (growth inhibition, followed by cell death at high doses). Belinostat is an HDAC inhibitor that causes a coincident response (partial growth inhibition and death activation at every dose, but with varied proportions across doses). Using a conventional analysis of these data, all three drugs can be observed to substantially decrease the relative viability of U2OS cells (Figure 2G). Although Palbociclib is noticeably less potent, it is not clear from these data that Palbociclib fails to activate cell death. Using a GR-based analysis, which reports how drugs affect the net population growth rate, it can be more accurately interpreted that Palbociclib-treated populations continue to expand at all doses, whereas high doses of Camptothecin or Belinostat result in population shrinkage, and thus, cell death must have been activated following high doses of these drugs (Figure 2H). However, a GR-based analysis fails to account for varied coordination between growth and death, which can lead to erroneous conclusions about the level of cell death activated by a given drug. For instance, based on the GR data, one might conclude that 1 µM Camptothecin activates cell death to a greater extent than 1 µM Belinostat, given that Camptothecin results in a GR value of -0.25 and Belinostat results in a GR value of 0; a -0.25 GR value means the population is shrinking 25% as fast as the untreated population is expanding. Using a GRADE-based visualization and analysis, it can be observed that Camptothecin and Belinostat differ in their growth/death coordination. Thus, at the 1 µM dose, both Belinostat activates death at a higher rate than Camptothecin (0.8% death/hr for Belinostat, compared to 0.7% death/hr for Camptothecin), but the Camptothecin-treated population shrinks faster due to a more pronounced growth inhibition induced by this drug (Figure 2I).
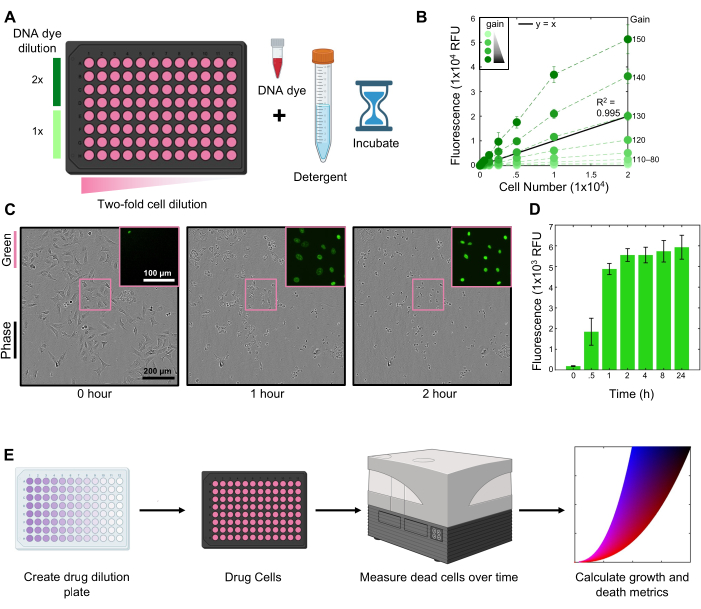
Figure 1: Flow chart of the FLICK protocol. (A) Create a cell dilution to optimize the DNA dye concentration and linearity of fluorescence measurements on a microplate reader. (B) Gain titration across cell number for 2 µM SYTOX Green in U2OS cells. A gain of 130 provides the most linear signal across all cell numbers. (C) Phase images of U2OS cells lysed with 1.5% Triton-X over time using a 10x objective. The boxed region is a zoomed-in view of SYTOX Green fluorescence. Scale bars are representative of all images. (D) SYTOX Green signal following the lysis at the indicated times. Data are mean +/- SD for 3 biological replicates. (E) A conceptual overview of the assay, from set up through analysis. Key steps are the creation of a drug dilution plate, drugging of cells in a multi-well plate, acquisition of fluorescence measurements over time, and calculation of growth and death metrics. For panels B and D, data are mean ± SD for n = 3 independent biological replicates. Please click here to view a larger version of this figure.
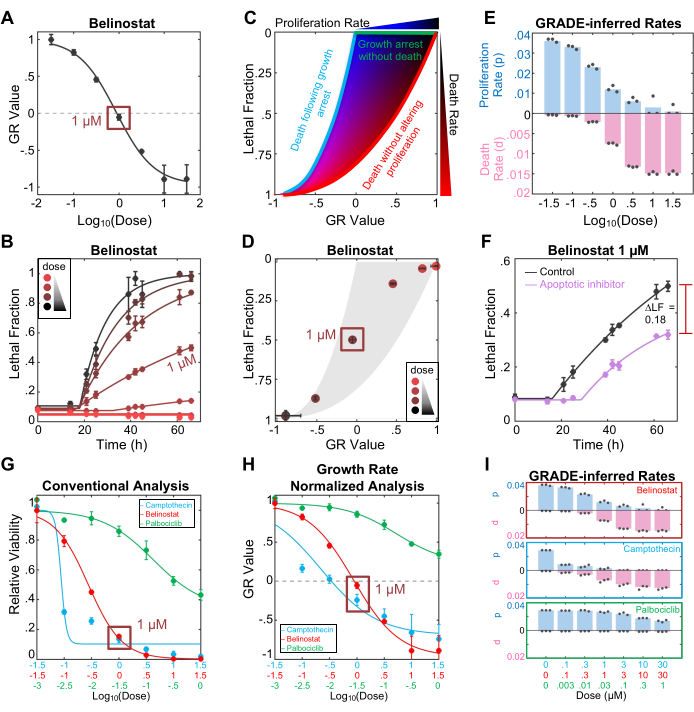
Figure 2: Characterizing drug responses using the FLICK assay and GRADE-based analysis. (A) GR metric for the HDAC inhibitor, Belinostat, over a dose of doses. The data shown are from the assay endpoint, which was 66 h. At 1 µM, Belinostat has a GR value of approximately 0, indicating a cytostatic response. (B) Lethal fraction kinetics for Belinostat. At 1 µM Belinostat induces nearly 50% lethality, which is not captured by the GR value. (C) GRADE phase diagram based on a simulation of all possible growth and death rates. Simulation was based on the doubling time of U2OS in this assay (27.12 h). (D) GRADE plot for Belinostat across dose at assay endpoint. (E) Bar plot of GRADE-inferred growth (p, population doublings per hour) and death (d, fraction of population that dies per hour) rates for doses of Belinostat. Rates were determined from simulated rates in (C) and data from (D). (F) Lethal fraction plot for 1 µM Belinostat treated with (purple) or without (black) 50 µM of the apoptotic inhibitor z-VAD. ΔLF = 0.18 was calculated by subtracting the average max LF from the inhibitor-treated condition from the average max LF from the control. (G) Relative viability (number of live cells in the drug-treated condition, divided by the number of live cells in the untreated condition) for three drugs following a 72 h exposure. All drugs reduce cell viability, but the contribution of cell death to the response is not known. (H) GR metric for three drugs in (G). These data demonstrate that two of the three drugs result in population shrinkage, indicated by GR < 0. The contribution of cell death is still unclear, although cell death must have been activated when the GR value is negative. (I) GRADE-inferred rates for three drugs. Palbociclib is non-lethal but results in growth arrest. Camptothecin is biphasic; lower doses result in growth arrest while higher doses result in growth arrest followed by lethality. Belinostat is a coincidental drug; the dose range cuts through the middle of the GRADE plot, indicating all doses result in a growth perturbation and cell death activation. Data in all panels are the mean ± SD for three independent biological replicate experiments. Please click here to view a larger version of this figure.
| Dye | Kinetic | End Point |
| SYTOX Green | Y | Y |
| CellTox Green | Y | Y |
| NucSpot 500/515, 594/615, 750/780 | Y | Y |
| YOYO-3 | Y | Y |
| Propidium Iodide | N | Y |
| 7-AAD | N | Y |
Table 1: DNA dyes and their application in FLICK. Many types of dyes can be used for end point-only FLICK assays. Fewer dyes meet the conditions to be incubated continuously throughout the assay. Other dyes, including dyes that are not green, have not been deeply characterized and should be carefully evaluated prior to use.
| Death Pathway | Inhibitor | Typical Dose |
| Intrinsic apoptosis | ZVAD-FMK | 50 µM |
| Extrinsic apoptosis | Z-IETD | 30 µM |
| Ferroptosis | Ferrostatin-1 | 10 µM |
| Necroptosis | Necrostatin-1 | 50 µM |
| Pyroptosis | VX-765 | 50 µM |
| Parthanatos | Rucaparib | 1 µM |
| Autophagic Dependent Cell Death | Hydroxychloroquine (HCQ) | 10 µM |
| Lysosomal Dependent Cell Death | E-64D | 10 µM |
| Cuproptosis | Tetrathiomolybdate (TTM) | 5 µM |
| Oxeiptosis | N-acetyl-l-cysteine (NAC) | 2 mM |
| MPT-driven necrosis | Cyclosporin A (CsA) | 10 µM |
Table 2: Cell death pathways and their inhibitors. Cell death inhibitors should be validated against a canonical activator for each death pathway. Selected inhibitor doses should ideally not affect cell viability.
Supplementary Table 1: GRADE calculator. Spreadsheet for calculating drug-induced growth and death rates from FLICK-based GR and FV data. Users should input GR and FV data, along with the length of the assay in hours, cell doubling time in hours, and starting/ending population sizes. Please click here to download this File.
Discussion
The FLICK assay is a robust method for generating a comprehensive evaluation of a drug's effect on the growth and death of a cell population. Because this method does not directly count cells, critical steps in the protocol are to ensure assay linearity and complete lysis during the triton permeabilization steps. The correct permeabilization time can be identified visually, as highlighted in this protocol, or quantitatively by reading plate fluorescence over time and identifying when the signal plateaus. In our experience, however, when in doubt, there does not appear to be any cost to waiting longer: for the DNA dyes that we have used in the FLICK assay, the signal intensity remains stable for several days following lysis. We regularly use the SYTOX Green nucleic acid stain, and while we have not validated each fluorophore referenced in Table 1, several other protocols demonstrate their effective use13,15,16. Assessing the linearity of a given cell-impermeant DNA dye across a range of cell numbers and instrument settings will assure robust and quantitatively reliable data.
Some cell lines are more difficult to lyse than others, and it should never be assumed that the lysis time for one cell line is the same as another13. For some cell lines, at higher densities, cell lysis causes the cells to lift off the well as a sheet, which can cause plate reader error. This can be avoided by reducing the starting seeding density or pipette-mixing the wells after lysis and before reading. To improve the lysis time or lysis efficiency, the Triton-X solution can be adjusted to a higher or lower percentage, but we have yet to encounter a scenario where this was necessary.
Experimental design and drug plating layout should be carefully considered when robust growth data is desired. Creating a drug dilution plate for the FLICK assay is the same as any other 96-well plate assay. However, including many controls covering different parts of a plate's landscape will assist in creating robust insights into growth kinetics and can help determine if systematic growth variation exists. Avoiding the outer wells of a plate will produce more precise growth data, as these wells are more sensitive to temperature, oxygen, or humidity fluctuations, affecting their growth. Lastly, using a pseudo-randomized drugging pattern, such as switching a drug's location on replicate plates, will provide a more accurate evaluation of drug behavior.
Some limitations exist for the accurate use of the FLICK assay. These limitations are mostly related to kinetic inference, as the endpoint data are experimentally observed values. Both the live cell proliferation kinetics and the death kinetics depend on some assumption regarding the growth trajectory for live cells over time in the drug-treated condition. In this protocol, we describe this growth trajectory as exponential and with a uniform rate over time. While these features are likely to be observed in the absence of a drug, they may not be adhered to in the presence of a drug. The FLICK method is accurate, not because the assumptions about growth trajectories are always correct, but instead, because the impact of any incorrect assumptions is minimized if the drug-treated population does not proliferate very much (i.e., less than 2 or 3 population doublings in the assay period). We have not encountered drugs that cannot be accurately profiled in FLICK; however, the theory would suggest that the FLICK assay will cease to be accurate for a drug in which death occurs very slowly, over long periods of time that are several multiples of the cell doubling time (note: we have not identified any drugs with these features3). However, the impact of any incorrect assumptions for the growth kinetics of the total population will not affect the death onset time obtained from the LED fit or the final maximum LF values, as these are constrained by the empirically measured dead cell values, and the final total cell values that were experimentally determined at the end of the assay.
Notwithstanding these limitations, the FLICK assay enables insights that are challenging to generate using other drug response methods. Most drug response methods generate a signal proportional to the number of live cells, and these methods can be used to quantify how drugs affect the net population growth rate (i.e., GR value or equivalent) but cannot accurately discriminate between the cytotoxic versus cytostatic effects of a drug. Alternatively, microscopy-based assays that measure both live and dead cells can certainly generate a comprehensive picture. However, microscopy-based assays may struggle to count dead cells after the cells decompose to debris, as will happen rapidly in the context of many non-apoptotic forms of cell death. A key feature of FLICK is that measurements are made in a plate reader, which aggregates the total dead cell fluorescence rather than directly counting dead cells. Thus, the dead cell signal in FLICK does not depend on the intact cellularity of dead cells, which, in the context of non-apoptotic cell death, is uniquely essential. Furthermore, a unique feature of the FLICK assay is the ability to measure both the live and dead cell populations using the same reagent. Thus, the FLICK assay produces measurements of live and dead cells with equal sensitivity. This feature enables analysis using the GRADE method and improves the accuracy of GRADE's simultaneous growth and death rate calculations. The included Supplementary Table 1 includes a template used for visualization of drug GRADE, where the user can input their assay length, calculated growth rate from step 7.1, starting and ending cell numbers or fluorescent readings from control conditions, and the calculated LF and GR from steps 5.4 and 6.3. The file contains a simulation of all pairwise combinations of 50 growth and death rates that will automatically update based on the user-defined parameters. The template generates a GRADE-based visualization of the drug response and the GRADE-inferred growth (p) and death (d) rates.
Future work should explore the application of the FLICK assay in 3D cell culture scenarios or in the evaluation of drug responses of tumor-derived organoids. Linearity and sensitivity of the FLICK data have not been deeply interrogated in these contexts, but in theory, the FLICK assay should be effective with some modifications to integrate fluorescence across 3-dimensional samples. Additionally, adding additional cell-specific labels would help to discriminate between two or more cell types in co-culture. These advances will be valuable for exploring immune cell interactions with cancer cells. Finally, assays like SPARKL inspire the use of death-specific reporters in the FLICK assay format, which may improve throughput while maintaining the accuracy of cell death insights11,16.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
We thank all past and present members of the Lee Lab for their contributions to our lab's perspective on evaluating drug responses. This work was supported by funding from the National Institutes of Health to MJL (R21CA294000 and R35GM152194).
Materials
| Name | Company | Catalog Number | Comments |
| Belinostat | ApexBio | A4612 | |
| Camptothecin | ApexBio | A2877 | |
| Conical centrifuge tube, 15mL | Fisher Scientific | 12-565-269 | |
| DMEM | Corning | 10017CV | For seeding and drugging cells |
| DMSO | Fisher Scientific | MT-25950CQC | For seeding and drugging cells |
| Fisherbrand 96-Well, Cell Culture-Treated, U-Shaped-Bottom Microplate | Fisher Scientific | FB012932 | For seeding and drugging cells (pin plate) |
| Greiner Bio-One CELLSTAR μClear 96-well, Cell Culture-Treated, Flat-Bottom Microplate | Greiner | 655090 | For seeding and drugging cells |
| IncuCyte S3 | Essen Biosciences | Any phase microscope will work | |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html | MATLAB version R2023b, a license is required |
| Microplate fluorescence reader | Tecan | Spark | For measuring dead cell fluorescence |
| Palbociclib | ApexBio | A8316 | |
| PBS | Corning | 21-040-CM | Any PBS works |
| Spark Multimode Microplate Reader | Tecan | https://www.tecan.com/spark-overview | SparkControl software version 2.2 |
| Sterile reservoir, 25 mL | Fisher Scientific | 13-681-508 | For seeding and drugging cells |
| Sytox Green nucleic acid stain | Thermo Fisher Scientific | S7020 | DNA stain for measuring dead cell fluorescence |
| Triton-X 100 | Thermo Fisher Scientific | J66624-AP | For permeabilizing cells |
| U2OS | ATCC | HTB-96 | An example cell line used in this protocol |
| Z-VAD-FMK | ApexBio | A1902 |
References
- Hafner, M., Niepel, M., Sorger, P. K. Alternative drug sensitivity metrics improve preclinical cancer pharmacogenomics. Nat Biotechnol. 35 (6), 500-502 (2017).
- Dixon, S. J., Lee, M. J. Quick tips for interpreting cell death experiments. Nat Cell Biol. 25 (12), 1720-1723 (2023).
- Schwartz, H. R., et al. Drug GRADE: An Integrated Analysis of Population Growth and Cell Death Reveals Drug-Specific and Cancer Subtype-Specific Response Profiles. Cell Rep. 31 (12), 107800(2020).
- Hafner, M., Niepel, M., Chung, M., Sorger, P. K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 13 (6), 1-11 (2016).
- Galluzzi, L., et al. Molecular mechanisms of cell death recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25 (3), 486-541 (2018).
- Tsvetkov, P., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 375 (6586), 1254-1261 (2022).
- Holze, C., et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat Immunol. 19 (2), 130-140 (2018).
- Maltese, W. A., Overmeyer, J. H. Methuosis Nonapoptotic Cell Death Associated with Vacuolization of Macropinosome and Endosome Compartments. Am J Pathol. 184 (6), 1630-1642 (2014).
- Inde, Z., Forcina, G. C., Denton, K., Dixon, S. J. Kinetic Heterogeneity of Cancer Cell Fractional Killing. Cell Rep. 32 (1), 107845(2020).
- Forcina, G. C., Conlon, M., Wells, A., Cao, J. Y., Dixon, S. J. Systematic Quantification of Population Cell Death Kinetics in Mammalian Cells. Cell Syst. 4 (6), 1-18 (2017).
- Gelles, J. D., et al. Single-Cell and Population-Level Analyses Using Real-Time Kinetic Labeling Couples Proliferation and Cell Death Mechanisms. Dev Cell. 51 (2), 277-291.e4 (2019).
- Richards, R., et al. Drug antagonism and single-agent dominance result from differences in death kinetics. Nat Chem Biol. 16 (7), 791-800 (2020).
- Richards, R., Honeywell, M. E., Lee, M. J. FLICK: an optimized plate reader-based assay to infer cell death kinetics. STAR Protoc. 2 (1), 100327(2021).
- Honeywell, M. E., et al. Functional genomic screens with death rate analyses reveal mechanisms of drug action. Nat Chem Biol. 20 (11), 1443-1452 (2024).
- Jannoo, R., Xia, Z., Row, P. E., Kanamarlapudi, V. Targeting of the Interleukin-13 Receptor (IL-13R)α2 Expressing Prostate Cancer by a Novel Hybrid Lytic Peptide. Biomolecules. 13 (2), 356(2023).
- Gelles, J. D., Chipuk, J. E. Why not add some SPARKL to your life (and death)! Mol Cell Oncol. 7 (1), 1685841(2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved