Method Article
Cell-Free Dot Blot as a Practical and Adaptable Immunoassay Platform for the Detection of Antibody Response in Human and Animal Sera
In This Article
Summary
We describe a recently developed immunoassay platform based on the principles of cell-free synthetic biology and the dot-blot technique for customizable detection of antibody response in human and animal sera.
Abstract
The string of global pathogenic outbreaks over the past two decades has highlighted the importance of serosurveillance strategies. Immunoassay platforms that serve to detect disease-specific antibodies in patients' sera are at the core of serosurveillance. Common examples include enzyme-linked immunosorbent assays and lateral flow assays; however, while these are gold standard methods, they require pathogen-specific consumables and specialized equipment, which limits their use outside of well-resourced laboratories.
We have recently developed a novel immunoassay platform called Cell-Free Dot-Blot (CFDB) and validated it using human and animal sera against SARS-CoV-2. Unlike conventional immunoassays, CFDB patient serum samples are immobilized to a solid phase (nitrocellulose membrane), while the target antigen is suspended in the mobile phase of the assay. To improve access to serosurveillance capabilities, CFDB antigens are produced on demand and with low-burden infrastructure using in vitro protein expression. Here, the antigen is fused with a peptide tag that can be detected using a single universal reporter protein for any CFDB assay. The result is that the CFDB does not require access to a multi-well plate reader or purified commercial molecular assay components. With these design considerations, CFDB addresses the shortcomings of existing immunoassay platforms by providing accessibility to non-centralized laboratories, adaptability for emerging pathogens, and affordability for lower-income communities.
In the current article, we will provide a step-by-step protocol to prepare and perform a CFDB immunoassay. Using our recent work on SARS-CoV-2 CFDB as an example, we will cover antigen DNA design for on-demand cell-free production, followed by preparation of the CFDB reporter protein, immobilization of serum samples on the solid phase, and finally, antigen-binding and detection steps of the assay. We anticipate that by following these instructions, researchers will be able to adapt the CFDB assay to detect immune responses in human and animal sera to any given pathogen.
Introduction
The COVID-19 pandemic revealed the critical need for affordable, scalable diagnostic tools, particularly for low-resource settings1. Conventional immunoassays like enzyme-linked immunosorbent assays (ELISAs) have proven essential for detecting immune responses2,3. However, their high cost, reliance on complex reagents, and dependence on specialized equipment limit their accessibility, especially during global health crises. In response to these challenges, we developed the Cell-Free Dot Blot (CFDB), a low-cost, adaptable immunoassay platform designed for the detection of anti-SARS-CoV-2 antibodies in human and animal sera.
CFDB leverages cell-free synthetic biology for the rapid, on-demand production of viral antigens using linear DNA templates4,5. This eliminates the need for traditional cell-based cloning, expression, and purification processes, significantly speeding up antigen production while reducing costs. The CFDB method simplifies antibody detection by using a dot blot format, where sera are directly spotted onto nitrocellulose membranes. This system obviates the need for expensive multi-well plates and specialized lab equipment, allowing for a simple "dipping" workflow for incubation and wash steps. The platform also utilizes a SpyCatcher-SpyTag system, where a SpyCatcher2-Apex2 peroxidase chimera acts as a universal secondary detection reagent5,6. This is produced using standard Escherichia coli-based expression, which eliminates dependence on costly commercial antibody conjugates. As a result, the CFDB system can perform serological assays with performance comparable to ELISAs at a significantly lower cost-about $3 USD per 96 sample assay compared to over $300 USD for a commercial ELISA kit5.
To demonstrate CFDB's effectiveness, we tested its ability to detect antibodies in precharacterized human and animal sera. Our results closely correlated with ELISA in identifying COVID-19-positive and -negative samples. In addition to human diagnostics, we evaluated CFDB's utility in animal models, testing sera from SARS-CoV-2-infected hamsters and those vaccinated with recombinant Nucleocapsid protein. These tests confirmed CFDB's potential for use in both human and veterinary diagnostics, making it a versatile tool for monitoring immune responses across species. One of the key advantages of CFDB is its flexibility. By simply modifying the DNA template encoding the antigen of interest, the platform can be rapidly adapted to detect antibodies against different pathogens, making it valuable for future pandemic preparedness. Its low cost, simple workflow, and minimal infrastructure requirements make it particularly suitable for decentralized laboratories and low-resource environments, where access to commercial diagnostics is limited.
In this work we will provide step-by-step instructions for preparing and conducting a CFDB assay. First, we cover the design and synthesis of linear DNA templates for cell-free production of antigens, which are the assay's primary detection reagents. We then describe steps for the preparation of the assay's secondary detection reagent SpyCatcher2-Apex2. After that, we provide instructions for the cell-free production and quality-check of antigens themselves. Finally, we describe in detail the process for conducting a CFDB assay on human or animal serum samples.
Protocol
All hamster experiments were performed at the National Microbiology Laboratory (NML) at the Public Health Agency of Canada, approved by the Canadian Science Centre for Human and Animal Health, and following the Canadian Council on Animal Care guidelines. All human serum/plasma samples were obtained commercially for in-house testing or provided by clinical collaborators to the NML for independent testing at the NML.
1. Design and preparation of antigen linear expression templates (LETs)
- Design the cell-free expression LETs for the target antigen based on the instructions in Norouzi et al.7 and containing an N/C-terminal SpyTag as described in Norouzi et al.5.
NOTE: Here we provide details for the preparation of SARS-CoV-2 Nucleocapsid Protein (NP, amino acids 2-419).- Ensure that the LET for the SARS-CoV-2 NP (Figure 1 and Supplemental File 1) contains 5' and 3' Ter sites with a 50 base pair buffer sequence before the T7 promoter and after the stop codon.
- Tag the NP protein on the N-terminus with His6-SpyTag-TEV.
- Obtain the protein sequence from the UniProt Database (Accession Code P0DTC9) and codon-optimize it for E. coli-based expression using the IDT codon optimization tool8. Order this sequence for commercial synthesis as a single-stranded DNA fragment resuspended in water at 10 ng/μL.
NOTE: The TEV protease cleavage site and the His6 tag are not essential features in the LET design.
- PCR amplify the DNA fragment with the universal Ter FW (GGCTCCGAATAAGTATGTTGTAACTAAAGTGCGGCC
ACGATGCGTCCGGCGTAGAGGATCG) and Ter RV (CCGAGGCAATAAGTATGTTGTAACTAAAGTGCTCAG
CTTCCTTTCGGGCTTTGTTAGCAGCC) primers using the high-fidelity DNA polymerase kit (see the Table of Materials)- Set up a 100 µL reaction as follows: 75 µL of nuclease-free water, 20 µL of 5x DNA polymerase buffer, 2 µL of 10 mM dNTPs (final concentration 200 µM), 0.5 µL of 100 µM Ter-FW primer, 0.5 µL of 100 µM Ter-RV primer, 1 µL of 10 ng/µL single-stranded LET, 1 µL(2U) of DNA polymerase.
- Use the following PCR settings: initial denaturation at 98 °C for 30 s; 35 cycles: 98 °C for 6 s, 60 °C for 15 s, 72 °C for 90 s; hold at 4 °C.
- Purify the PCR product using a commercial PCR purification kit, check its quality by running a sample on a 1% agarose gel, and measure its concentration on a UV-Vis spectrophotometer.
NOTE: Crude PCR products can also be used directly for cell-free expression. However, purification allows for a more standardized procedure.

Figure 1: Linear expression template for SARS-CoV-2-NP. A schematic representing features of the His-SpyTag-SARS-CoV-2 NP linear DNA template. Key DNA template elements are labeled. The coding sequence for the protein of interest, here the NP, is placed under the transcriptional control of a T7 promoter for efficient expression. At the N-terminus, the NP protein is appended with a SpyTag for specific detection using the SpyCatcher2-Apex2 detection reagent. The x6His-tag and TEV protease sites, although included as part of the general LET design, are dispensable for CFDB purposes. At the termini of the linear DNA template, "upstream" and "downstream" Ter sites, each preceded by respective 50 base pair buffer sequences, are included for Tus-mediated protection against exonucleolytic DNA degradation in the cell-free lysate. Abbreviations: NP = nucleocapsid protein; LET = linear expression template; TEV = tobacco etch virus; CFDB = cell-free dot blot. Please click here to view a larger version of this figure.
2. Purification of the SpyCatcher2-Apex2 reporter protein
- Use the pET24b-SpyCatcher2-Apex2 plasmid, originally constructed in Norouzi et al.5 (full sequence in Supplemental File 2 and plasmid map in Supplemental Figure S1) to prepare the SpyCatcher2-Apex-2 reporter protein.
- Prepare agar plates and lysogeny broth (LB) containing 50 μg/mL kanamycin. Transform E. coli BL21 (DE3) cells with the pET24b-SpyCatcher2-Apex2 plasmid. Inoculate a single colony into a 15 mL starter LB culture and grow overnight at 37 °C with shaking at 250 RPM.
- Next day, add 10 mL of the starter culture into 500 mL of fresh LB media containing 50 μg/mL kanamycin. Incubate at 37 °C with shaking at 250 RPM until the culture reaches an optical density at 600 nm of 0.6-0.8 (~3 h).
- Induce the expression of SpyCatcher2-Apex2 by supplementing the culture with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside and 1 mM 5-aminolevulinic acid hydrochloride. Reduce the growth temperature to 30 °C and allow the culture to incubate for another 4 h.
- Harvest the bacteria by centrifugation at 8,000 × g for 15 min. Proceed to cell lysis or alternatively store the pellet at -80 °C until use.
- Resuspend the pellet in 20 mL of lysis buffer containing 50 mM Tris-HCl (pH 7.8), 300 mM NaCl, 1 mg/mL lysozyme, EDTA-free protease inhibitor tablet, and 1 mM dithiothreitol (DTT). Lyse the cells by sonication at 50 % amplitude with 5 s ON and 10 s OFF intervals for a total ON time of 3 min.
- Clarify the lysate by centrifugation at 20,000 × g for 1 h at 4 °C. Pass the supernatant through a 0.2 μm syringe filter.
- Add hemin chloride to a final concentration of 250 μM to the clarified lysate and incubate overnight at 4 °C. Proceed to protein purification.
NOTE: Incubation with hemin chloride maximizes heme incorporation into the Apex-2 peroxidase for optimal enzyme activity. - Add 2.5 mL of Ni resin to the clarified lysate and incubate at 4 °C with gentle shaking for 45 min. Apply the mixture to a gravity flow column and wash the resin with 50 mL of Tris buffer (50 mM Tris-HCl (pH 7.8), 300 mM NaCl, and 1 mM DTT).
- Elute the SpyCatcher2-Apex2 protein in 25 mL of Tris buffer containing 400 mM imidazole. Concentrate and buffer exchange the eluate into Tris buffer using a centrifugal filter unit, aiming for a final volume of 0.5-1.0 mL.
- Use the molar extinction coefficient of SpyCatcher2-Apex2 (27,390 M-1 cm-1) to determine the protein concentration on a UV-Vis spectrophotometer. Add glycerol to a final concentration of 40% and store the aliquots at -20 °C.
NOTE: This protocol is expected to yield approximately 40 mg of highly pure and active SpyCatcher2-Apex2, sufficient for 400 CFDB runs on a standard 96-sample blot size. - For detection of SpyTagged proteins using conventional western blot9 and for CFDB experiments, always block membranes in 5% non-fat dry milk in 1x Tris-buffered saline containing 0.05% Tween-20 (TBST). Use the SpyCatcher2-Apex2 protein at a final concentration of 10 μg/mL in blocking solution.
NOTE: The SpyCatcher2-Apex2 reporter requires enhanced chemiluminescence (ECL) solution for signal development. The ECL solution can be obtained commercially or prepared in-house according to instructions in Mruk et al.10. The final ECL solution here consists of 0.4 mM p-coumaric acid, 2.5 mM luminol, and 0.015% H2O2 in 100 mM Tris-HCl (pH 8.6).
3. Cell-free production and quality check of antigens
- Prepare and assemble an E. coli BL21 cell-free expression lysate and reaction components by following the instructions in Levine et al.11 and Norouzi et al.5. Supplement the final reaction mixture with 5 μM Tus protein and 1.2 μM T7 RNA Polymerase as follows for a representative 100 μL reaction: 14.6 µL of Solution A, 14 µL of Solution B, 33.3 µL of E. coli BL21 lysate, 2.5 µL of 200 mM Tus protein, 1.2 µL of 100 mM T7 RNA polymerase, 1 µL of 1.5 µM linear DNA template, 33.4 µL of nuclease-free water.
NOTE: See Supplemental File 3 for key instructions on preparing an E. coli BL21 cell-free lysate and Supplemental Table S1 for a detailed recipe of Solutions A and B. The E. coli cell-free expression system can also be prepared using alternative protocols or obtained commercially, as long as the reaction is supplemented with Tus and T7 RNA Polymerase. - Perform an initial 5 μL-scale expression test by adding 10% (v/v) crude PCR product to the cell-free reaction in a PCR tube. Incubate without shaking at 30 °C for 15 h.
- Check the expression quality of the NP antigen by loading 1 μL of the cell-free reaction on a 12% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and subsequently transfer it to a nitrocellulose membrane for western blot as described in step 2.12. Use the SpyCatcher2-Apex2 reporter protein for blot labeling via the SpyTag, and optionally, a specific commercial antibody (here an anti-SARS-CoV-2-NP) against the target antigen.
- Assemble a 1 mL-scale cell-free reaction with 15 nM purified LET product to be used for the CFDB assay. Incubate the expression mixture in a 15 mL conical tube with shaking at 80 RPM for 15 h at 30 °C. Check the expression quality using western blot and store 50 μL aliquots at -20 °C.
4. Serum samples
- Obtain human/animal serum (or plasma) samples from commercial sources or clinical collaborators or in-house using standard procedures and following guidelines from institutional human research ethics committee or animal care and use committee. Ensure that appropriate sample pretreatment measures have been taken to mitigate risks of contamination.
NOTE: Pretreatment measures may include solvent-detergent treatment, heat inactivation, and testing for bloodborne virus markers. A useful protocol for serum and plasma isolation from whole blood is available12. As with other immunoassay platforms, serum samples collected early (<3 weeks) after initial infection or disease onset are not expected to contain adequate levels of an antibody response and their results should be treated with caution. - For quality verification of the SARS-CoV-2 NP CFDB reagents, use The National Institute for Biological Standards and Control (NIBSC) World Health Organization (WHO) international reference panel for anti-SARS-CoV-2 immunoglobulin, which contains x1 pre-COVID-19 and x4 SARS-CoV-2-positive samples with varying levels of anti-NP immunoglobulins13. Alternatively, use precharacterized healthy and positive serum samples from other sources.
- For CFDB experiments, obtain or prepare a negative control sample by pooling multiple (>3) healthy serum samples to increase the accuracy and facilitate the analysis and interpretation of results.
5. Cell-free Dot Blot (CFDB) procedure
- Download and print the master-grid image file (Supplemental Figure S2). The master grid is a 6 x 6 cm pattern containing 12 x 12 circles of 2 mm diameter each and provides a spotting capacity similar to a 96-well plate.
- Sandwich the grid firmly between two layers of adhesive PCR plate sealing film and cut to size along the outer grid borders. Use a 2 mm biopsy punch to hollow each marked circle.
NOTE: This master grid can be reused multiple times after wiping with 70% ethanol. - Cut a 6.5 x 6.5 cm piece of nitrocellulose membrane and position it under the master grid on a clean surface, securing the setup using adhesive tape as shown in Figure 2 and Supplemental Figure S3. Use a marker pen to mark the outermost circle positions on the nitrocellulose membrane to be used as a guide to cut the membrane after sample spotting.
- Dilute the serum samples 1/10 in 1x phosphate-buffered saline (pH 7.4). Use a micropipette to dispense triplicate 0.4 μL volumes of each sample onto the nitrocellulose membrane at predetermined grid positions. It will take approximately 15 s per spot to dispense.
NOTE: Be sure to include both negative and positive control samples with each assay, as this will be required for the analysis of results. - Allow 10 min at ambient temperature for the spotted samples to bind and dry. Using tweezers, carefully retrieve the nitrocellulose membrane and cut along the marked outer circles.
- Block the membrane in 10 mL of blocking solution (5% non-fat dry milk in TBST) in a 10 cm Petri dish for 30 min at room temperature, shaking gently at 100 RPM.
- Thaw and add a 50 μL aliquot of the cell-free antigen expression mixture to 5 mL of blocking solution in a 10 cm Petri dish. Transfer the membrane directly to this antigen-containing solution and incubate at room temperature for 1 h, shaking at 100 RPM.
NOTE: This will be the primary detection step where the SpyTagged antigen will bind to antibody-containing spot positions, if present. - Rinse the membrane, wash for 5 min, and rinse again in TBST before proceeding to the secondary detection step.
- Incubate the membrane in 10 mL of blocking buffer containing 10 μg/mL of purified SpyCatcher2-Apex2 protein, for 1 h while shaking at 100 RPM.
- Rinse the membrane and wash 2 x 5 min in TBST, with a final rinse in TBS.
- Stick and secure a piece of parafilm on a clean work area close to the blot imaging instrument. Remove excess liquid by tapping the membrane using tweezers and place the membrane on top of the parafilm.
- Immediately add 3 mL (100 μL/cm2) of ECL solution on top of the membrane and incubate at room temperature for exactly 90 s.
- Immediately tap-dry the membrane and transfer to a chemiluminescence-compatible imaging instrument for visualization of results.
NOTE: Optimal imaging time can vary on different instruments. It is recommended to use the instrument's default auto-acquisition time settings, and if absent, use precharacterized serum samples to optimize image acquisition time. - Use the instrument's image analysis function to obtain spot intensities, including for three blank (background) positions on the nitrocellulose membrane; maintain a constant measurement volume per spot. Export the data to a spreadsheet, remembering to correctly label each spot position.
- Calculate the Mean and Standard Deviation (SD) of triplicate spot intensities. Then, subtract the nitrocellulose membrane background from all samples.
- Use the following equation to obtain a cut-off value for the interpretation of -/+ results:
(Mean of negative controls) + (3 x SD of the negative controls). Consider samples positive if their average intensity falls above the cut-off value and negative if their average intensity falls below the cut-off value.
NOTE: A schematic of the CFDB procedure is provided in Figure 3.
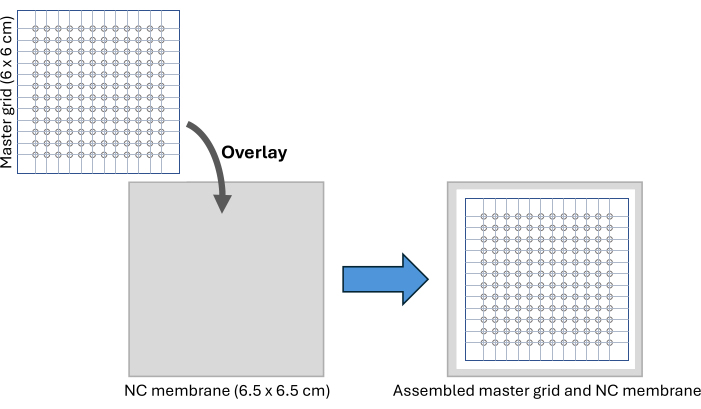
Figure 2: CFDB assembly. A schematic of the CFDB master grid and NC membrane assembly setup. The master grid is overlaid on the NC membrane to provide a regular, addressable pattern for spotting and immobilization of serum samples. Abbreviations: CFDB = cell-free dot blot; NC = nitrocellulose. Please click here to view a larger version of this figure.

Figure 3: A schematic representation of the CFDB workflow. In a CFDB assay, a small amount (<0.4 μL) of 10x diluted serum samples is manually dispensed onto a precut nitrocellulose membrane (left panel) in discrete, addressable locations (middle panel). Depositing one serum sample per spot in triplicate spots and immobilizing the protein content, including the sera's total antibody reservoir, on the solid NC substrate (beige spots in the middle panel). In this example, anti-NP antibodies contained in the serum samples can be first bound by the CFDB primary detection reagent SpyTag-NP and finally detected by the CFDB secondary detection reagent SpyCatcher2-Apex2 (right panel-magnified bubble). This figure was taken from Norouzi et al.5. Abbreviations: CFDB = cell-free dot blot; NP = nucleocapsid protein; LET = linear expression template. Please click here to view a larger version of this figure.
Results
PCR amplification of the linear expression template for target antigen
To PCR-amplify the SARS-CoV-2 NP LET, universal Ter forward and reverse primers were used as described in protocol section 1.2 and 1 μL of the product was checked on an agarose gel (Figure 4) before proceeding to the purification of the PCR product.

Figure 4: PCR of linear expression template. A 1% agarose gel showing in Lane 1: 1 kb DNA Ladder and Lane 2: the PCR-amplified linear DNA template for the His-SpyTag-SARS-CoV-2 NP (1,630 bp). Abbreviations: PCR = polymerase chain reaction; NP = nucleocapsid protein. Please click here to view a larger version of this figure.
Purification of the SpyCatcher2-Apex2 reporter protein
The SpyCatcher2-Apex2 protein was prepared as described in protocol section 2. To ensure adequate heme incorporation into the Apex2 peroxidase, 5-aminolevulinic acid hydrochloride (a heme precursor) and hemin chloride were added during protein synthesis and after cell lysis, respectively. Samples from all purification steps were loaded on an SDS-PAGE gel to ensure the quality of the final product (Figure 5).
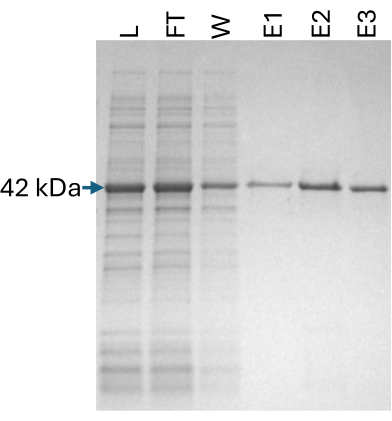
Figure 5: SpyCatcher2-Apex2 purification. A 12% SDS-PAGE gel showing fractions from the purification steps of the SpyCatcher2-Apex2 protein. L: lysate (2 μL), FT: flowthrough (2 μL), W: wash (10 μL), E1-3: elution 1-3 (10 μL) each. The expected band at 42 kDa is marked. Abbreviation: SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis. Please click here to view a larger version of this figure.
Cell-free expression and quality verification of the target antigen
After performing a 5 μL-scale pilot expression of the NP protein and quality check using SDS-PAGE, a 1 mL-scale expression was conducted as described in protocol section 3. A 1 μL sample of the final cell-free expression mixture was subjected to western blot separately for detection by SpyCatcher2-Apex2 and an anti-NP commercial antibody (Figure 6). This step ensures that the antigen is expressed successfully and that it can bind specifically to both anti-NP immunoglobulins and the SpyCatcher2-Apex2 reporter protein.
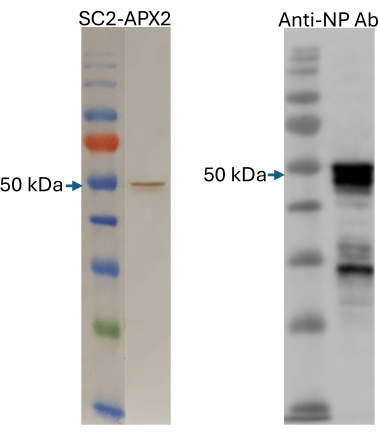
Figure 6: Quality check of cell-free produced antigens. Western blot detection of the cell-free produced NP protein using SpyCatcher2-Apex2 (left panel) and an anti-NP rabbit polyclonal antibody (right panel). The expected full-length product at 50 kDa is marked. The size marker in both gels is the Color Pre-stained Protein Standard, Broad Range. This figure was adapted from Norouzi et al.5. Abbreviation: NP = nucleocapsid protein. Please click here to view a larger version of this figure.
Verification of CFDB reagents using precharacterized human sera
To confirm that the CFDB reagents are of high quality and that the CFDB protocols are implemented successfully, we used a precharacterized serum panel provided by the NISBC-the WHO international reference panel for anti-SARS-CoV-2 immunoglobulin 20/26813. This panel consists of x1 pre-COVID-19 (negative) and x4 SARS-CoV-2-positive samples with varying levels of antibody response (Supplemental Table S2). We also acquired a serum panel from RayBiotech (CoV-PosSet) containing 10 pre-COVID-19 (negative) and 20 presumed SARS-CoV-2-positive samples. We pooled equal fractions from all 10 negative sera to use as a negative control across all experiments performed in our laboratory. This pooling strategy increases the accuracy and robustness of the results as single negative samples may exhibit subtle levels of background signal that may complicate the interpretation of results.
A CFDB assay was performed on the precharacterized WHO reference panel and RayBiotech's CoV-PosSet according to protocol section 5 and the blot was imaged. The volume tool in Image Lab Software was used to draw uniform circles on each spot position and export individual signal intensities. After subtracting NC membrane background intensity from all samples, the cut-off value ((Mean of negative controls) + (3 x SD of the negative controls)) was also subtracted from the average signal intensities. Samples are considered anti-NP positive if their signal intensity falls above the cut-off value. An image of the blot and corresponding signal intensity chart is presented in Figure 7. The results demonstrate that using a cell-free produced SARS-CoV-2 NP protein, the CFDB technique can correctly identify all negative samples and all positive samples, except two samples from the RayBiotech set (RB P5-collected 34 days post symptom onset (dpso) and RB P10-collected 4 dpso). Further inspection of the supplier's datasheet confirmed that both these samples also appear negative for anti-S1RBD (Spike Receptor Binding Domain) IgG by ELISA and anti-N IgG/M by lateral flow assay (Supplemental Table S3), suggesting a lack of detectable antibody levels in these samples.

Figure 7: Verification of CFDB using precharacterized commercial human sera. (A) Blot image representing an anti-SARS-CoV-2 NP CFDB on the WHO international reference panel and RayBiotech PosSet serum samples. Samples were spotted in triplicate column-wise and identified by labels. (B) Graph representing the quantified chemiluminescence signal intensity values for the blot in panel A. The negative control was used to set the cutoff value, and all measurements are means of triplicate spots ± standard deviations (SD). This figure was taken from Norouzi et al.5. Abbreviations: Neg Cont = negative-control sample obtained by pooling all 10 preCOVID-19 samples from RayBiotech; RB N1 to N10 = RayBiotech presumed-negative samples 1 to 10; RB P1 to P20 = RayBiotech presumed positive samples 1 to 20; CFDB = cell-free dot blot; NP = nucleocapsid protein; WHO = World Health Organization. Please click here to view a larger version of this figure.
Independent testing of the CFDB assay using human patient sera
To demonstrate the applicability of the CFDB assay, we provided CFDB materials and instructions to an independent collaborating laboratory in a different location (National Microbiology Laboratory (NML), Winnipeg). The aim was to independently test the platform using human patient samples collected in the clinic by the field studies group at the NML. In total, 24 samples (x12 negative and x12 positive-confirmed by ELISA for NP antibodies (Supplemental Table S4) were subjected to CFDB and the results were analyzed as described above. As seen in Figure 8, CFDB correctly identified all x12 negative and x12 positive samples, confirming the robustness and practicality of the assay in an independent test.

Figure 8: Independent testing of CFDB using human patient sera at the NML. (A) Blot image representing an anti-SARS-CoV-2 NP CFDB on 12 COVID-19-negative (N1 to N12) or 12 positive (P1 to P12) patient samples. Sera were spotted in triplicates column-wise, as labeled. (B) Quantified chemiluminescence signal intensity values from the blot in panel A. All measurements are means of triplicate spots ± standard deviations (SD). This figure was taken from Norouzi et al.5. Abbreviations: CFDB = cell-free dot blot; NML = national microbiology laboratory; NP = nucleocapsid protein. Please click here to view a larger version of this figure.
Independent testing of the CFDB assay using SARS-CoV-2 infected hamster sera
Using the same CFDB reagents and instructions, the NML team then performed a CFDB assay on SARS-CoV-2 infected hamster sera to test whether this assay can also be used to detect antibody response in animal samples. In total, 36 samples were tested, x12 preinfection and 24 postinfection sera. The presumed positive sera were from animals infected with 105 50% tissue culture infective doses (TCID50) of SARS-CoV-2 that were sampled on 140 days-post infection (dpi 140, n = 3); or reinfected on dpi 140 and sampled 5 days later (dpi 5, n = 21). As shown in Figure 9, all preinfection sera had signal intensities below the cut-off value and 18 out of 24 post infection (all from 5 days post reinfection) sera elicited discrete positive signal intensities. Interestingly, of the six presumed positive samples missed by CFDB, three samples (all from the initial dpi 140) were also undetectable using a standard anti-NP ELISA (Supplemental Table S5), indicating a loss or decrease in antibody levels over time.
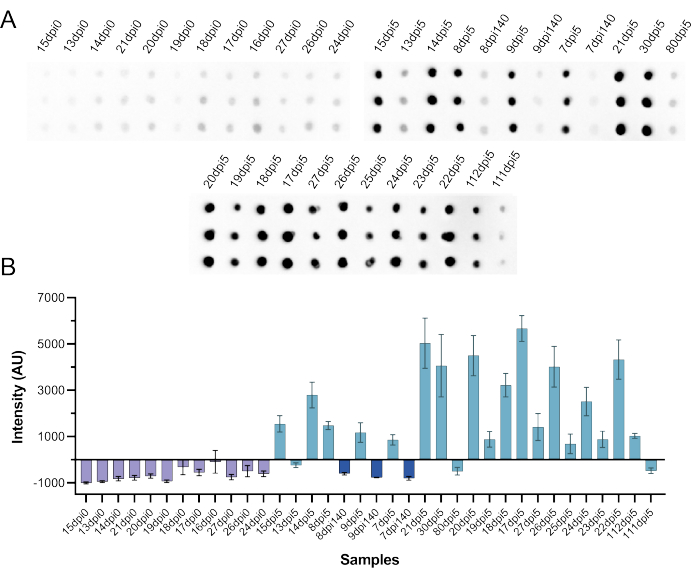
Figure 9: Independent testing of CFDB using SARS-CoV-2 infected hamster sera at the NML. (A) Blot image presenting an anti-SARS-CoV-2 NP CFDB on hamster sera preinfection (dpi 0, n = 12), 140 days after infection (dpi 140, n = 3), and 5 days after re-infection (dpi 5, n = 21) with 105 TCID50 of SARS-CoV-2. Each sample was deposited as triplicate spots column-wise and labeled according to the scheme in the original NML study. (B) Quantified chemiluminescence signal intensity values from the blot in panel A. All measurements are means of triplicate spots ± standard deviations (SD). This figure was taken from Norouzi et al.5. Abbreviations: CFDB = cell-free dot blot; NML = national microbiology laboratory; NP = nucleocapsid protein; dpi = days post infection; TCID50 = 50% tissue culture infectious dose. Please click here to view a larger version of this figure.
Supplemental File 1: Nucleotide sequence for the His-SpyTag-SARS-CoV-2 NP linear expression template. Abbreviation: NP = nucleocapsid protein. Please click here to download this File.
Supplemental File 2: Full nucleotide sequence of the pET24b SpyCatcher2-Apex2 plasmid. Please click here to download this File.
Supplemental File 3: An overview of the key steps in preparation of an E. coli BL21 cell-free protein expression lysate. Please click here to download this File.
Supplemental Figure S1: Construct map for the pET24b SpyCatcher2-Apex2 plasmid. Please click here to download this File.
Supplemental Figure S2: A printable copy of the CFDB master grid. Abbreviation: CFDB = cell-free dot blot. Please click here to download this File.
Supplemental Figure S3: An image of the CFDB NC membrane and master grid assembly setup. The NC membrane is sandwiched between a glass plate on the bottom and the master grid pattern on the top, securing the assembly using stationary tape. This figure was taken from Norouzi et al.5. Abbreviations: CFDB = cell-free dot blot; NC = nitrocellulose Please click here to download this File.
Supplemental Table S1: Complete recipes for Solution A and Solution B components of the cell-free reaction mixture. Please click here to download this File.
Supplemental Table S2: Sample Information for NIBSC WHO Reference Panel 20/268 (also see Figure 7). Abbreviations: NISBC = The National Institute for Biological Standards and Control; WHO = World Health Organization. Please click here to download this File.
Supplemental Table S3: Sample Information RayBiotech CoV-PosSet (also see Figure 7). Please click here to download this File.
Supplemental Table S4: NP ELISA results for human patient serum samples (performed at NML). See also Figure 8. Abbreviations: NP = nucleocapsid protein; ELISA = enzyme-linked immunosorbent assay; NML = national microbiology laboratory. Please click here to download this File.
Supplemental Table S5: NP ELISA results for SARS-CoV-2 infected hamster sera (performed at NML). See also Figure 9. Abbreviations: NP = nucleocapsid protein; ELISA = enzyme-linked immunosorbent assay; NML = national microbiology laboratory. Please click here to download this File.
Discussion
COVID-19 highlighted the importance of accessible and robust diagnostics for controlling infection outbreaks and optimizing global health strategies. Serological testing that detects protective antibodies proved essential for tracking transmissibility patterns of new variants, identifying hot spots, guiding vaccine development, triaging suspected cases and protecting vulnerable populations14. The pandemic also exposed inequities in testing accessibility, exasperated by backlogs and requirements for facilities with sophisticated equipment15. The focus is thus now on strengthening our global response to future outbreaks, which will require new innovations that reduce testing barriers and can be rapidly adapted for unpredictable threats.
CFDB is a paper-based immunoassay that simplifies serological testing, conventionally performed by ELISAs or lateral flow assays, by immobilizing patient samples in the solid phase and delivering mobile-phase cell-free produced antigens for detection of anti-viral antibodies5. The assay can be conducted in basic lab environments with western blot capabilities. Meanwhile, reagents for CFDB can be produced locally in basic research settings and readily interchanged to repurpose the assay for new pathogens. The chimeric SpyCatcher2-Apex2 universal detection reagent is an enabling feature that obviates the need for sourcing commercial secondary antibody conjugates and is produced efficiently in an E. coli expression system. CFDB also provides high throughput capabilities that parallel clinical assays at a fraction of the cost, at only $3 USD per assay kit compared to roughly $300 USD for a commercial 96-well ELISA.
Configuring the CFDB for COVID-19 serological testing was achieved by designing linear DNA templates that instructed the cell-free transcription-translation system to produce the target antigen, SARS-CoV-2 NP. Human and animal sera were then spotted onto nitrocellulose membrane grids and incubations were performed with the cell-free antigen expression mixture followed by the SpyCatcher2-Apex2 reporter protein. In this study, CFDB was performed on sera from COVID-19 negative and positive subjects, both within our laboratory and in an independent setting (NML), and overall, a good correlation with ELISA was achieved for distinguishing positive versus negative samples. By using pooled healthy sera as the CFDB negative control and calculating an assay cut-off value based on this sample, the risk of false negative/positive identification was minimized.
High sensitivity of the CFDB assay is manifested by its ability to clearly distinguish all samples in the extensively precharacterized NIBSC's WHO Reference Panel 20/26813, particularly the sample with the lowest antibody titer (Sample 20/140 low, see Supplemental Table S2 and Figure 7). Similarly, CFDB's high specificity is demonstrated by the lack of false-positive identification for any of the 35 healthy sera tested in this work. Importantly, multi-site clinical validation confirmed that the CFDB protocol is sufficiently straightforward for widespread use and maintains high performance, comparable to standard ELISAs, when executed by different users. In our experience and as shown by others16, the cell-free antigens produced in E. coli lysates are stable for at least 3 months at -20 °C, and multiple cell-free antigen batch preparations perform indistinguishably from one another.
Several considerations should be accounted for prior to adopting the CFDB platform widely and pivoting the system design towards new pathogens. The CFDB platform does require that the viral antigen or antigen fragment is properly synthesized by a cell-free expression system so that antigenic moieties are present and available for antibody binding. For example, we initially intended to also include the SARS-CoV-2 Spike receptor binding domain (RBD) in our CFDB assays. However, despite a high yield expression and because of its requirement for complex disulfide linkages (unlike the NP antigen that lacks Cysteine amino acids), the RBD product readily aggregated and appeared insoluble in the E. coli extract, precluding its use as a reliable CFDB probe. In such scenarios where complex posttranslational modifications are required for proper folding and antigenic function, the use of eukaryotic-based cell-free systems is recommended. As with other peroxidase-based immunoassays, it is also imperative that serum samples with visible hemolysis are excluded due to the potential for hemoglobin peroxidase activity confounding the results. Finally, the CFDB is designed to detect the total antibody response and does not report on the specific Immunoglobulin class involved in antibody-antigen interactions.
In conclusion, the CFDB is a novel diagnostic technique that merges elements of cell-free synthetic biology and dot blotting to streamline serological testing. The CFDB offers a cost-effective alternative to conventional ELISAs that can be readily deployed in research laboratories worldwide when additional testing capacity is needed, as is often the case during a pandemic. The CFDB can thus serve as a frontline tool in the next global pandemic response, which can be rapidly adapted for emerging threats and will strengthen the testing capacities of low and middle-income countries.
Disclosures
M.N. and K.P. are co-inventors of the cell-free dot blot method. A provisional patent application related to this work has been filed (PCT/CA2024/050097, filed January 2024).
Acknowledgements
S.S. and R.Z. are supported by funding from the Defense Advanced Research Projects Agency (DARPA), Contract No. N66001-23-2-4042. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government. This work was supported by funds to K.P. from the CIHR Foundation grant program (201610FDN-375469), CIHR Canada Research Chair Program (950-231075 and 950-233107), University of Toronto's Medicine by Design Initiative, which receives funding from the Canada First Research Excellence Fund and funds to K.P., from Defense Research and Development Canada's, Canadian Safety and Security Program (contract 39903-200137). Figure 1 and Supplemental Figure S1 were created using SnapGene Viewer.
Materials
| Name | Company | Catalog Number | Comments |
| 1 kb DNA ladder | NEB | N3232 | Used as a size marker for agarose gels |
| 20 Amino acids | Sigma-Aldrich | LAA21-1KT | A component of the cell-free reaction Solution B |
| 2 mm biopsy punch | Integra Miltex | 33-31-P/25 | For preparation of CFDB master grid |
| 5-aminolevulinic acid hydrochloride | Sigma-Aldrich | A3785 | Heme precursor for induction of SpyCatcher2-Apex2 |
| Agarose powder | BioShop | AGA002 | For electrophoretic analysis of DNA |
| Anti-SARS-CoV2-Nucleocapsid antibody | Sinobiological | 40588-T62 | For WB detection of cell-free produced NP antigen |
| Bio-Rad ChemiDoc XRS+ | Bio-Rad | N/A | Gel imager instrument |
| Centrifugal concentrators | Cytiva | 28-9323-60 | For concentration and buffer exchange of proteins |
| Centrifuge | Eppendorf | EP022628257 | For harvesting bacterial culture |
| Coenzyme A sodium salt hydrate (CoA) | Sigma-Aldrich | C3144 | A component of the cell-free reaction Solution A |
| Color pre-stained protein standard (broad range) | NEB | P7719 | Used as a size marker for SDS-PAGE gels |
| Covid-19 serum panel | RayBiotech | CoV-PosSet | Pre-characterised sera for verification and optimisation of CFDB reagents/conditions |
| D-(−)-3-Phosphoglyceric acid disodium salt (3-PGA) | Sigma-Aldrich | P8877 | A component of the cell-free reaction Solution B |
| Dithiothreitol (DTT) | Sigma-Aldrich | 10197777001 | Component of buffers |
| E. coli 5-alpha | NEB | C2987 | for plasmid preparation |
| E. coli BL21 | NEB | C2530 | For preparation of cell-free lysates |
| E. coli BL21 (DE3) | NEB | C2527 | For expression of SpyCatcher2-Apex2 |
| EDTA-free protease inhibitor tablet | Sigma-Aldrich | 11836153001 | A component of E. coli lysis buffer |
| Eppendorf New Brunswick Innova 43/43R Incubator Shaker | Eppendorf | EPM1320 | Incubator for growing E. coli cells for protein expression and cell-free lysate preparation |
| Folinic acid | Sigma-Aldrich | 47612 | A component of the cell-free reaction Solution A |
| Glycerol | Sigma-Aldrich | G9012 | Component of SpyCatcher2-Apex2 storage buffer |
| Hemin chloride | Sigma-Aldrich | H9039 | For Heme supplementation of SpyCatcher2-Apex2 |
| Hydrogen peroxide 30% | Sigma-Aldrich | H1009 | For making ECL reagent |
| Image Lab Software | Bio-Rad | 1709690 | Software for ChemiDoc gel imaging instrument |
| Isopropyl-b-D-1-thiogalactopyranoside | Bioshop | IPT001 | For induction of SpyCatcher2-Apex2 expressikon |
| Kanamycin Sulfate | Sigma-Aldrich | 60615 | For preparation of SpyCatcher2-Apex2 bacterial culture |
| LB agar | BioShop | LBL406 | For E. coli growth |
| LB broth | BioShop | LBL407 | For E. coli growth |
| luminol | Sigma-Aldrich | A4685 | For making ECL reagent |
| Lysozyme | Sigma-Aldrich | L6876 | For lysis of bacterial cells |
| Magnesium Acetate | Sigma-Aldrich | M5661 | A component of the cell-free reaction Solution B |
| NEBExpress Ni resin | NEB | S1428S | For purification of SpyCatcher2-Apex2 |
| Non-fat dry milk | Bioshop | SKI400 | For blocking of WB and CFDB membranes |
| Parafilm | Bemis | 2099-1337410 | For ECL-incubation of CFDB blots |
| p-coumaric acid | Sigma-Aldrich | C9008 | For making ECL reagent |
| pET24b-SpyCatcher2-Apex2 plasmid | Pardee Laboratory | N/A | Used for the expression of SpyCatcher2-Apex2 protein |
| Petri dish | Fisherbrand | FB0875712 | Container for WB and CFDB membrane incubation |
| Phosphate buffered saline 10% | BioShop | PBS405 | Component of buffers |
| Potassium Glutamate | Sigma-Aldrich | G1501 | A component of the cell-free reaction Solution B |
| Potassium Oxalate Monohydrate (Oxalic acid) | Sigma-Aldrich | 223425 | A component of the cell-free reaction Solution A |
| Precast SDS-PAGE gel | BioRad | 4561036EDU | For electrophoretic analysis of protein |
| Q5 high-fidelity DNA polymerase | NEB | M0491 | For PCR amplification of LETs |
| QIAquick PCR purification kit | Qiagen | 28106 | For purification of LETs |
| Ribonucleotide Solution Set | NEB | N0450S | A component of the cell-free reaction Solution A |
| Skim milk powder | BioShop | SKI400 | Used as a blocking agent for western blot and CFDB |
| Sodium Chloride | Sigma-Aldrich | S9625 | A component of protein purification buffers |
| Sonicator | Qsonica | Q500 | For lysis of bacterial cells |
| Spermidine | Sigma-Aldrich | S2626 | A component of the cell-free reaction Solution A |
| Syringe filter | Sigma-Aldrich | SLGSR33SS | For sterilisation of buffers and solutions |
| Thermocycler | BioRad | T100 | Instrument for incubating PCR reactions |
| Transfer RNA (tRNA) | Sigma-Aldrich | R8759 | A component of the cell-free reaction Solution A |
| Tris buffered saline | Thermo Scientific | J60764.K2 | Component of WB and CFDB wash buffer |
| Trizma base | Sigma-Aldrich | T1503 | For making tris buffer |
| Tunair SS-5012 Half-Baffle Shake Flask, 2.5 L | Cole-Parmer | RK-01835-39 | Vessel for growing E. coli cells for protein expression and cell-free lysate preparation |
| Tween-20 | BioShop | TWN510 | Component of WB and CFDB wash buffer |
| Tweezer | Almedic | 7728-A10-100 | For handling CFDB NC membrane |
| UV-Vis spectrophotometer | Thermo Scientific | 13400518 | Used for the measurement of nucleic acid and DNA concentration and bacterial culture OD |
| WHO International Reference Panel for anti-SARS-CoV-2 immunoglubulin | NISBC | 20/268 | Pre-characterised sera for verification and optimisation of CFDB reagents/conditions |
| β-Nicotinamide adenine dinucleotide hydrate (NAD) | Sigma-Aldrich | 10127965001 | A component of the cell-free reaction Solution A |
References
- Carter, L. J., et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci. 6, 591-605 (2020).
- Amanat, F., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 26 (7), 1033-1036 (2020).
- Stadlbauer, D., et al. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 57 (1), e100(2020).
- Pardee, K., et al. Portable, on-demand biomolecular manufacturing. Cell. 167 (1), 248-259.e12 (2016).
- Norouzi, M., et al. Cell-free dot blot: an ultra-low-cost and practical immunoassay platform for detection of anti-SARS-CoV-2 antibodies in human and animal sera. Microbiol Spectr. 11 (2), e0245722(2023).
- Lam, S. S., et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 12 (1), 51-54 (2014).
- Norouzi, M., Panfilov, S., Pardee, K. High-efficiency protection of linear DNA in cell-free extracts from Escherichia coli and Vibrio natriegens. ACS Synth Biol. 10 (7), 1615-1624 (2021).
- IDT codon optimization tool. , Integrated DNA Technologies. https://www.idtdna.com/pages/tools/codon-optimization-tool (2025).
- General protocol for western blotting. , Bio-Rad. https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6376.pdf (2025).
- Mruk, D. D., Cheng, C. Y. Enhanced chemiluminescence (ECL) for routine immunoblotting. Spermatogenesis. 1, 121-122 (2011).
- Levine, M. Z., Gregorio, N. E., Jewett, M. C., Watts, K. R., Oza, J. P. Escherichia coli-based cell-free protein synthesis: Protocols for a robust, flexible, and accessible platform technology. J Vis Exp. (144), (2019).
- Plasma and serum preparation. , ThermoFisher Scientific. https://www.thermofisher.com/ca/en/home/references/protocols/cell-and-tissue-analysis/elisa-protocol/elisa-sample-preparation-protocols/plasma-and-serum-preparation.html (2025).
- Mattiuzzo, G., et al. WHO/BS.2020.2403 Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 antibody. , (2020).
- Gong, F., Wei, H., Li, Q., Liu, L., Li, B. Evaluation and comparison of serological methods for COVID-19 diagnosis. Front Mol Biosci. 8, 682405(2021).
- Peeling, R. W., et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 20 (9), e245-e249 (2020).
- Smith, M. T., Berkheimer, S. D., Werner, C. J., Bundy, B. C. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques. 56 (4), 186-193 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved