Method Article
Identification and Classification of Position-specific GABAA Receptor Subunit Missense Variants for Their Role In Hippocampal Pyramidal Neurons
In This Article
Summary
This study introduces a multiscale framework, spanning from DNA to protein function and neural behavior. It presents a novel approach for investigating predicted pathogenic mutations in the GABAA receptor subunit, hypothesizing that epileptogenic mutations and proximal mutations, predicted as pathogenic, may produce similar effects on the CA1 pyramidal neuron model.
Abstract
Understanding the effects of functionally unknown variants in epilepsy associated genes is crucial for elucidating disease pathophysiology and developing personalized therapeutics. With a multiscale framework, spanning from DNA sequence to protein function and neural behavior, we describe a novel approach for predicting and investigating pathogenic mutations, hypothesizing that epileptogenic mutations in the GABAA receptor subunit and nearby predicted mutations may produce similar effects on the CA1 pyramidal neuron model. By exploring the characteristic relationships between predicted pathogenic mutations and proximal epileptogenic mutations, the study aims to estimate the effects of predicted mutations based on the effects of epileptogenic mutations on hippocampal pyramidal neuron simulations.
The methodology begins with the collection of GABAA receptor γ2 subunit genetic data, followed by data cleaning and formatting performed in R using a custom script. Next, ensemble predictors will be applied to identify and prioritize the pathogenic missense variants of the γ2 subunit. Mapping a specific pathogenic variant (predicted) to the subunit structural domains shared by epileptogenic mutations will be illustrated, accompanied by molecular modeling of their effects and consideration of evolutionary conservation. Then, variant-specific meta-analysis and parameter normalization will be performed, followed by correlation analysis to identify any significant relationships between predicted mutations and proximal epileptogenic mutations. Using a Python-based neural simulator, multi-compartmental conductance-based neuron model, reflecting the effect of wild-type and epileptogenic mutants will be described. Simulation of neural responses generated by epileptogenic GABAA receptor subtype will be considered for the rough estimation of the predicted pathogenic variants' effect on neural response. To our knowledge, this is the first protocol exploring a multiscale framework to estimate the effects of GABAA receptor variants on neuronal behavior, crucial for epilepsy research. This protocol can serve as a foundation for enhancing predictions of cellular phenotypes caused by potentially pathogenic variants of GABAA receptors associated with epilepsy.
Introduction
For nearly all human diseases, genetic variation plays a significant role in individual susceptibility. Therefore, understanding how sequence variations relate to disease risk offers a valuable way to uncover key processes involved in disease development and identify new approaches for prevention and treatment1. This also applies to neurodevelopmental disorders, which rank among the most prevalent chronic medical conditions in pediatric primary care2. Conditions such as autism spectrum disorder, intellectual disability, and epilepsy illustrate how genetic variation significantly influences individual susceptibility during development3.
The developing brain is more susceptible to epileptic seizures than the adult brain due to genetically programmed neurodevelopmental mismatch in the critical balance between excitation and inhibition4. As GABA (gamma-aminobutyric acid), the primary inhibitory neurotransmitter in the adult brain, is excitatory during embryonic and early postnatal development, this is not favorable to the stability needed to prevent seizures in young brains. This temporary state, caused by the lack of sufficient expression of K-Cl co-transporters5, can contribute to an increased risk of seizure activity in the presence of dysfunctional GABAA receptors. GABAA receptors mediate excitatory and inhibitory actions of GABA, depending on the intracellular concentration of the Cl- ion6. Thus, as the brain matures, mutations in the GABAA receptor-encoding genes, as well as in other ion channels, distort excitability, and mutations in genes involved in neuronal metabolism, cell signaling, and synapse formation7, can cause conditions like childhood absence epilepsy8.
Clinical interventions are increasingly leveraging genetic analysis to improve precision in treating neurodevelopmental disorders2. Genetic testing in pediatric epilepsy presents potential targets for precision medicine approaches9, highlighting the significance of genetic variants in guiding treatment decisions. In addition, ~25% of epilepsy patients with de novo mutations receive genetic diagnoses that identify potential targets for precision medicine, underscoring the significant value of genetic variants in guiding treatment decisions10. This has been fueled by advancements in next-generation sequencing technologies, such as targeted gene panels, whole-exome sequencing, and whole-genome sequencing, which have dramatically accelerated genetic discoveries11. However, the increasing number of new gene discoveries comes with a challenge when results yield a variant of unknown significance (VUS), a classification that reflects conflicting evidence or insufficient information regarding the variant's molecular role in disease pathogenesis. Variants classified as VUS correspond to one category within the five-tier variant classification system proposed by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP)12.
Addressing the challenge of functionally unknown genetic variants requires efforts across two key dimensions: clinical practice and research. Clinically, the uncertainty surrounding VUS can complicate patient management and decision-making13. From a scientific research perspective, identifying pathogenic variants among the increasing number of variants of uncertain significance and determining their roles in disease pathophysiology and phenotypic effects are crucial1. One ideal scenario would involve accurately predicting the molecular, neuronal, and network-level effects of all functionally uncharacterized variants, thereby minimizing the resources, time, and effort required for laboratory-based investigations. These aspects underscore the importance of accurately classifying genetic variants to enable precise diagnosis of genetic epilepsies, support personalized treatment, and facilitate the discovery of potential pharmacological targets. Current predictive tools14,15,16,17 are relatively accurate but typically provide only binary classifications (pathogenic vs. benign) and lack disease-specific insights into molecular pathophysiology, phenotypic consequences, and underlying mechanisms. Focusing on the unknown missense variants of selected GABAA receptor subunit-encoding genes, this paper presents a framework aimed at enhancing research guidance by incorporating contextual factors of variants such as molecular, evolutionary, and structural aspects, as well as simulations of neural pathology derived from in vitro biophysical data of epilepsy-associated mutations. Our methodology addresses the identification of unknown pathogenic variants of the γ2 subunit of the GABAA receptor, a key subunit involved in the pathophysiology of epilepsy18,19,20. This is followed by the exploration of position-specific matching of these predicted variants with the epilepsy-associated mutations characterized by structural and electrophysiological data. These data are then used to estimate the variant effect on a model of hippocampal pyramidal neuron expressing a GABAA receptor subtype, composed of γ2, α1, and β3 subunits (γ2-GABAA receptors), responsible for fast synaptic inhibition6. It is important to note that GABAA receptors assemble from a large subunit pool (α1-α6, β1-β3, γ1-γ3, δ, Ε, θ, π, and ρ1-ρ3) and depending on the subunit composition, GABAA receptors differ in their modulation, biophysical characteristics, as well as regional, cellular, and subcellular expression patterns coupled with specific functions6,21,22,23,24,25. Thus, the present study focuses on the γ2-GABAA receptors or γ2-containing GABAA receptors only.
GABAA receptor subunits are composed of characteristic structural features-a long N-terminal extracellular domain (ECD), four transmembrane spanning domains (TM1 to TM4), an intracellular linker connecting the TM1 and TM2, an extracellular linker connecting the TM2 and TM3, a large intracellular loop between TM3 and TM4 (TM3-TM4 loop), and a short extracellular C terminus6,26,27. It is suggested that the GABAA receptor functions via a complex "lock and pull" mechanism, where GABA binding locks the β and α subunits, causing them to pull on the extracellular domains (ECDs) of the subunits, rotating them counterclockwise27. This movement bends the transmembrane domains (TMDs), thereby opening the ion channel27. Thus, the channel activity appears to be coordinated together with structural cassettes within the GABAA receptors. It turns out that epilepsy mutations cause dysfunction in channel activity via distortion of these structural cassettes28. Consequently, our study is based on the idea that predicted pathogenic variants in proximity to functionally identified epileptogenic mutations in the specific structural cassettes of the GABAA receptor subunits may exhibit similar patterns of electrophysiological or biophysical distortion in channel function, as observed in cases of these epileptogenic mutations. While the presence of epileptogenic structural cassettes in the GABAA receptor subunits28 indirectly supports this notion, our study demonstrates the complexity and challenge of correlating biophysical parameters of epileptogenic mutations with those of predicted pathogenic mutations. To unmask these complex relationships, our framework is significant as it highlights a multiscale approach ranging from DNA to protein function and neural behavior critical for epilepsy research. This approach integrates computational genetics with molecular modeling and neural simulations while also emphasizing the importance of complementary methods, such as machine learning trained on large datasets, that could capture the effects of mutations on channel structure, activity, and neural excitability. In addition, the simulation of epileptogenic γ2-GABAA receptor activity on the hippocampal pyramidal neuron model allows the replication of in vitro cellular phenotype associated with GABAA receptor channelopathy and the demonstration of altered single-neuron responses at the center of network dysfunction.
Protocol
1. In silico prediction of pathogenic variants
- Variant data collection
- Using the ClinVar database29, search for variants of uncertain significance (VUS) in the coding region of the gene of interest via the website: https://www.ncbi.nlm.nih.gov/clinvar/. Enter the gene symbol (e.g., GABRG2) in the search bar and filter the results to include only the desired types of variants, such as single-nucleotide, missense variants with uncertain significance. Download and save the data as data.xlxs (Supplementary File 4: Supplementary Table S1). Record the date of the downloaded data.
NOTE: In the present protocol, human γ2 subunit of GABAA receptor, specifically Homo sapiens gamma-aminobutyric acid type A receptor subunit gamma2 (GABRG2), transcript variant 1, mRNA (NCBI Ref. seq.: NM_198904.4), also known as γ2L, will be analyzed. It is important to record the reference transcript of the gene of interest as well as other corresponding identifiers across different databases (UniProt, ENSEMBL, PDB) since different computational methods may require different identifiers (Supplementary File 4: Supplementary Table S2). In case the database or computational tool does not recognize the version numbers of the sequence identifiers, try both the ID with the version number (NM_198904.4) and without the version number (NM_198904). - Reference protein basic information
- In the NCBI database https://www.ncbi.nlm.nih.gov/, select Nucleotide in the search options and enter the NCBI Ref. seq. ID of the gene of interest (NM_198904.4). Then, by scrolling down on the right column, click on the Protein under the category Related information to find the protein (NP_944494.1) encoded by the transcript NM_198904.4. Using the information given for the protein NP_944494.1, record the sequence positions of the specific regions in the form of a table (Supplementary File 4: Supplementary Table S3).
NOTE: It is important to determine the preliminary known information for sequence position of functionally and structurally critical regions, motifs, or residues such as protein domains, phosphorylation sites, ligand binding sites, and molecular interaction interfaces. This can be achieved by combining database (NCBI, ENSEMBL, UniProt...) and literature searches.
- In the NCBI database https://www.ncbi.nlm.nih.gov/, select Nucleotide in the search options and enter the NCBI Ref. seq. ID of the gene of interest (NM_198904.4). Then, by scrolling down on the right column, click on the Protein under the category Related information to find the protein (NP_944494.1) encoded by the transcript NM_198904.4. Using the information given for the protein NP_944494.1, record the sequence positions of the specific regions in the form of a table (Supplementary File 4: Supplementary Table S3).
- Using the ClinVar database29, search for variants of uncertain significance (VUS) in the coding region of the gene of interest via the website: https://www.ncbi.nlm.nih.gov/clinvar/. Enter the gene symbol (e.g., GABRG2) in the search bar and filter the results to include only the desired types of variants, such as single-nucleotide, missense variants with uncertain significance. Download and save the data as data.xlxs (Supplementary File 4: Supplementary Table S1). Record the date of the downloaded data.
- Variant data organization
- Organize the data to meet the input requirements for the chosen predictors. Ensure that the format of the data retrieved is organized to match the requirements of the dbNSFP server http://database.liulab.science/dbNSFP. To do this, remove unnecessary columns from the data.xlsx file (Supplementary File 4: Supplementary Table S1 from step 1.1.1), keeping only the following columns in the specified order:
"GRCh38Chromosome", "GRCh38Location", "Name", "Protein change". - Save the file under a new file name: "data1.xlsx" (Supplementary Table S4). Format the data1.xlsx file in R by running the code (Supplementary File 1: Data_GABAA.R), which will save the formatted data as data1_output.xlsx (Supplementary File 4: Supplementary Table S5) in the working directory relevant to the R project.
NOTE: Different computational methods require different data types and formats. Collecting and organizing data according to specific format requirements, even for a dozen variants, can be prone to errors and time-consuming, so this step is important unless the variant pool is made up of only a few variants. Then, manual data organization may be possible.
- Organize the data to meet the input requirements for the chosen predictors. Ensure that the format of the data retrieved is organized to match the requirements of the dbNSFP server http://database.liulab.science/dbNSFP. To do this, remove unnecessary columns from the data.xlsx file (Supplementary File 4: Supplementary Table S1 from step 1.1.1), keeping only the following columns in the specified order:
- Pathogenicity prediction
- Transfer the content of the data1_output.xlsx file into the academic version of dbNSFP server30,31 accessed via http://database.liulab.science/dbNSFP. To do this, copy/paste or directly upload the file in .txt format.
- Ensure that the following options are preselected and confirmed in the server: HG38 (genome build), ClinPred32, and BayesDEL33 before submission. Within a few minutes, the server will generate the results.
NOTE: In the present protocol, two ensemble predictors, namely BayesDEL33 and ClinPred32, were selected for high accuracy34 and practicality. However, other predictors, such as AlphaMissense, which is available in the dbNSFP database30,31 can also be selected. The selection of in silico tools depends on several factors, including the generation of sufficient multiple lines of computational evidence for powerful prediction12. Ensemble predictors integrating the analysis of multiple predictive algorithms can serve this purpose. - Download the output file (a .txt format) and save it as data2.xlsx (Supplementary File 4: Supplementary Table S6).
- Set the filters in data2.xlsx (Supplementary File 4: Supplementary Table S6) by clicking on the filter option in the menu and determining the consensus variants in both columns by filtering for D. This will give the list of the most pathogenic variants; save it (see Consensus tab in Supplementary Table S6 [ Supplementary File 4]).
- Variant selection
- Among the consensus pathogenic predictions, determine the variants in the proximity of epileptogenic mutations obtained from the literature. Ensure that the latter have structural and biophysical parameters suitable for neuron modeling.
NOTE: This step is exploratory and is also related to surveying the protein of interest in terms of its structural, physicochemical, and biophysical parameters. In the present study, these data were obtained from Brünger et al.35 and Guo et al.36 in addition to a survey of epilepsy associated mutations. Besides, as an option, AlphaMissense37 scores were accessed from the dbNSFP database30,31 repeating step 1.3 (Supplementary File 4: Supplementary Table S7). More details are given in protocol sections 2.1.1 and 2.1.2 and in the Results (see "Clustering Variants for Structural and Biophysical Parameters"). - For basic visualization, use Protter38 ((https://wlab.ethz.ch/protter/start/), and HOPE39 (https://www3.cmbi.umcn.nl/hope/) servers to examine the variants in the previous step in the context of selected GABRG2 gene mutations: P302L40 and K328M (or K289M41, when excluding the 39-residue signal peptide).
NOTE: Due to the enormous complexity, structural evaluation of variant effects should be conducted at multiple levels of analysis. Tools like Protter38 will allow the clear visualization of the variants in the context of topological features of the protein and user-friendly servers such as HOPE39 will give insight into the variant effect by molecular modeling. In addition, a comprehensive literature review of the protein of interest is critical to identify and integrate the information about mutations associated with epilepsy. - Analysis of evolutionary conservation and structural insights
- Open Jalview42,43,44, an open-source program for editing, visualization, and analysis of proteins.
- Import sequences for alignment. Click on File in the top menu | Fetch sequences; select the database in the dialog box (such as UniProt); click on the retrieve IDs tab; and as described in the dialog box, enter UniProt accession IDs of the gene of interest (GABRG2) from human and other vertebrate species: P18507, P22723, Q6PW52, A0A2I3TKX0, F1RR72, A0A8I3MDZ2, A0A8M1P4D6. Click OK.
NOTE: UniProt accession numbers of proteins encoded by GABRG2 are as follows: P18507 (P18507-2) for Homo sapiens, P22723 for Mus musculus, A0A2I3TKX0 for Pan troglodytes, F1RR72 for Sus scrofa, A0A8I3MDZ2 for Canis familiaris, and A0A8M1P4D6 for Danio rerio. - Depending on the gene of interest, some sequences may not be annotated; therefore, perform a BLAST search to identify relevant information and potential homologs for a better contextual understanding. In this case, upload the FASTA format of protein sequences via the Add Sequences/From text box option under the File menu to produce multiple sequence alignments of the desired sequences.
- Once the alignment is loaded, observe the sequences displayed for multiple sequence comparison. Each row represents a sequence, and each column represents a position in the alignment. To determine the best alignment method, use different approaches; for instance, click on the Web services in the Sequence menu and select the Run T-Coffee with preset option, which allows optimal alignment.
- Right-click on the sequence P18507 Homo sapiens (the reference sequence in the present study) and set it as the reference sequence. Choose Format in the upper menu and click on Wrap for the visualization of the full alignment in the screen. In the same Format menu, click on the scale above to enhance the visualization of specific residue numbers. To further enhance visualization, adjust the color schemes by going to Color and selecting different options (e.g., Clustal Color, Chemical Property); modify the font size if required.
- Click on Calculate in the Menu bar and select Autocalculate consensus to highlight conserved regions.
- Focus on the position of variants of interest identified in the in-silico prediction step and examine specific variant positions. Annotate specific residues by right-clicking on them and selecting Add Annotation. Write the label (e.g., variant ID) with the appropriate color code and save.
NOTE: In the present analysis, P302L (purple) and A303T (red) were selected to visualize them in the multiple sequence alignment together with the structural data (see the next section).
- Three-dimensional reconstruction of the full protein showing the selected conserved residues
- In the file obtained from the previous step, right-click on the reference sequence (GABRG2 human) and select 3D structure data.
- Identify the appropriate structural data (7QNE, Chain C)26 from the dropdown menu and select Open new structure view with Jmol.
NOTE: This will allow the incorporation of the residues selected in the multiple sequence alignment into the structural data by Jmol, an open-source Java-based viewer for 3D chemical structures.
- Among the consensus pathogenic predictions, determine the variants in the proximity of epileptogenic mutations obtained from the literature. Ensure that the latter have structural and biophysical parameters suitable for neuron modeling.
2. Parameter selection and biophysical modeling
- Variant-specific meta-analysis and parameter normalization
- Survey current literature to gather identified subunit variants with electrophysiological data-channel conductance (gGABAA), deactivation time (τdeactivation), rise time (τrise), and maximum current amplitude (Imax). Provide the subunit compositions, cell type, and wild-type measurements for each case. Label the variants and their controls accordingly (e.g., known for variants with identified biophysical characteristics and known control for the wild-type measurements for each variant).
- Obtain AlphaMissense pathogenicity scores for variants with identified biophysical characteristics.
NOTE: See protocol section 1.3 for more details. - Create a data frame with subunit and amino acid position for each variant, the original and altered amino acids, pathogenicity score, and biophysical parameters obtained from the literature. To avoid experimental discrepancies, normalize the biophysical parameters for identified variants as x-fold changes on wild-type measurements.
- Comparative variant analysis by structural and functional characteristics
- Organize the predicted variants on a data frame; label accordingly (e.g., predicted for variants without literature available on their biophysical characteristics).
- Classify the variants by their location in the amino acid sequence and tertiary structure. Add structural classification parameters (e.g., localization in alpha helices, coils, beta sheets, extracellular, intracellular or transmembrane domains, pore lining, agonist binding, protein-protein interactions) on the data frame and provide information for each variant with respect to their amino acid position.
- Classify the variants by their distance to the membrane center and pore axis. Add distance to the pore axis and distance to the membrane center parameters on the data frame.
- Analyze the correlation among structural and biophysical parameters over known variants. If possible, evaluate the predicted variants with respect to the obtained correlations.
- Synapse and neuron model construction
- Use the Brian245, an open-source neural simulator developed in Python for modeling and simulating spiking neural networks, to build a multi-compartmental biophysical model of GABAergic synapse on a multi-compartmental conductance-based hippocampal pyramidal neuron.
- Design the conductance-based model by defining ion channel gating kinetics, passive and active parameters, and postsynaptic conductances. Define the conductance-based model as given in Supplementary File 2, which describes the equations used in the model.
- Set membrane capacitance (Cm) as 1 µF/cm2 and intracellular resistance (Ra) as 200 Ω.cm.
- Use the modified Hodgkin-Huxley type conductances for hippocampal pyramidal neurons39 with gL= 0.0003 S/cm2, gK= 0.036 S/cm2, EL = -76.5 mV, ENa = 50 mV, and EK = -90 mV.
- Adjust the density distribution of NaV channels over gNa as 0.05 S/cm2 for soma, 0.5 S/cm2 for axon initial segment (AIS) and node of Ranvier (NR), and 0.005 S/cm2 for dendrites. Set gK and gNa as 0 in myelinated segments.
- Build ion channel gating kinetics for NaV and KV as described in Supplementary File 2.
- Introduce synaptic currents (Isyn) as the summation of all glutamatergic and GABAergic synapses in a compartment. Include both fast AMPA receptor-mediated current (IAMPA) and slow NMDA receptor-mediated current (INMDA) in the glutamatergic current (Iglu). Include only fast GABAA receptor-mediated current in GABAergic current (IGABA). Assume that a constant amount of glutamate is released to the synapse for every presynaptic spike; therefore, the activation of receptors are spike-time-dependent (sAMPA and sNMDA) and the total receptor conductances (gAMPA and gNMDA) reflect the amount of glutamate that is released by every event.
- Use the synaptic model as described in Supplementary File 2.
NOTE: For a detailed explanation of the equations, see Supplementary File 2 describing the equations used in the model.
- Obtain the experimentally measured diameter for soma and neurites and the length of each neurite compartment and branching patterns from previous literature46,47. Reduce the real neuron morphology into a multi-compartmental model, by dividing the cell into multiple compartments, that accurately preserves the main branching structure and maintains bilateral symmetry.
- Set the morphological (segment length and diameter; i.e., d_soma: 30 µm; l_AH: 5 µm; d_AH_i: 1.5 µm; d_AH_f: 1.3 µm; l_AIS: 40 µm; d_axon: 1 µm; l_myseg: 100 µm; l_NR: 2 µm; l_AxTer: 4 µm; d_AxTer: 2 µm; l_approx: 100 µm; l_apmed: 100 µm; l_apdis: 200 µm; d_approx_i: 4 µm; d_approx_f: 3 µm; d_apmed : 2 µm; d_apdis: 2 µm; l_apLM: 70 µm; d_apLM: 2 µm; l_nAcDbasal: 400 µm; d_nAcDbasal: 1.4 µm; l_nAcDbasal_stem: 20 µm; d_nAcDbasal_stem: 1.5 µm) and biophysical parameters (as given in section 2.3.2) for each compartment of the pyramidal neuron model46,47 as also detailed in the Python script (Supplementary File 3: GABAAvar.py).
- Determine the biophysical parameters for the GABAergic synapse model by evaluating the wild-type control measurements obtained in step 2.1.1.
- Design the topology of the neuron model and assign morphological and biophysical parameters, which includes specifying the spatial arrangement and interconnections of the compartments, based on the previously obtained morphological and branching information. Assign the appropriate morphological (e.g., segment length and diameter) and biophysical parameters (Section 2.3.2) to each compartment of the model, as outlined in Supplementary File 3: GABAAvar.py.
- Building of synapses and current injection
- Create the presynaptic activity using SpikeGeneratorGroup (a class from the Brian2 library) as given in "GABAAvar.py" (Supplementary File 3). Connect the spike generator to the target compartment of the model neuron using Synapses class to model synaptic connections.
- Set a sustained constant current (Iinj) as 0.85 nA and place at the soma to mimic the subthreshold activity driven by baseline ionic current load at a given time as outlined in Supplementary File 3: GABAAvar.py.
- To build recording monitors, record voltage traces from target compartments using StateMonitor.
- Build and run the network.
- Build the network with the model neuron, connections, and monitors using Network.
- Set the time step of the simulation by defaultclock.dt (e.g., 0.01 ms).
- Run the simulation on the network with network.run(T*ms), where T is set as 1,000 ms in the example.
- Testing the impact of GABAA receptor missense mutations
- Define the impact of each missense mutation on channel kinetics through the biophysical parameters collected in step 2.1.1.
- Run the stimulation by altering these parameters and plot the results using "matplotlib.pyplot" as given in "GABAAvar.py" (Supplementary File 3).
- Test parameter combinations to analyze the changes in firing patterns and rates. Plot results for comparisons.
Results
This study utilizes a multiscale approach to predict and characterize the pathogenic variants in the γ2 subunit of the GABAA receptor, a key component in the pathophysiology of epilepsy. Through the use of predictive models, molecular modeling, evolutionary conservation, structural examination, correlation analysis, and neural simulations, this approach enhances the classification of variants, with significant relevance for epilepsy research and possibly for clinical use. The overall summary of the methodology is presented in Figure 1.
Comparative evaluation of two adjacent γ2 subunit mutations
Building on our assumption that predicted pathogenic mutations adjacent to epileptogenic mutations in GABAA receptor subunits may produce similar electrophysiological effects on channel function and neural behavior, we first conducted a brief examination of the relationship between a well-known epileptogenic mutation and a proximal predicted mutation of γ2 subunit.
Among the variants predicted as pathogenic (Supplementary Table S6), A303T (rs1581439874, ClinVar Accession: VCV000663033.6) is selected as an example. In addition to prediction by ensemble models, pathogenicity of A303T was confirmed by AlphaMissense scores (Supplementary File 4: Supplementary Table S7). A303T is in the second transmembrane domain of the GABAA receptor γ2 subunit and located next to the epileptogenic mutation P302L40, as shown in Figure 2. As assessed by molecular modeling, both γ2P302L and γ2A303T substitutions resulted in amino acids that have larger side chains, as shown in Figure 3. Both of the mutant and the wild-type residues are nonpolar in γ2P302L mutation, while in the γ2A303T, the mutant residue has a polar side chain and the wild-type residue has a nonpolar side chain. Both P302 and Ala303 are located in the subunit interaction interface with the β3 subunit (observed in 7QNB and 7QNA, respectively). Both P302 and Ala303 have comparable solvent-accessible surface area (SASA). In addition, both residues are 100% conserved across the span of vertebrate evolution (Figure 4, top panel). They are both located in the proximity of the second transmembrane region (TM2 domain) of the γ2 subunit, as shown in yellow in the three-dimensional reconstruction of the GABAA receptor protein (7QNE26, where A303, shown in red, is the first residue in this domain (Figure 4). Based on these comparable features and using a pyramidal neuron model, the simulation of proximal epileptogenic mutation(s) such as γ2 subunit mutation P302L40 may be used for the preliminary characterization of the predicted variant's (γ2A303T) effect on the neural response. In the next step, we expanded our analysis to a broader set of variants within the GABAA receptor subunits.
Clustering variants for structural and biophysical parameters
Following the comparative evaluation of two adjacent mutations in the previous section, we implemented a systematic approach to assess whether shared molecular features among variants could be identified. This phase aimed to explore whether consistent patterns emerge in structural, physicochemical, and biophysical features across the amino acids and variants, thereby providing further support for our initial hypothesis.
The data frame used in this study and the references are provided in Supplementary File 4: Supplementary Table S840, 48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63, Supplementary TableS936, and Supplementary TableS1035. In addition, correlations among structural and biophysical parameters were determined for each subunit and for all variants without subunit distinction (Supplementary File 4: Supplementary Table S11, Supplementary Table S12, Supplementary Table S13, Supplementary Table S14, and Supplementary Table S15). Information on structural parameters (localization on alpha helices, coils, beta sheets; extracellular, intracellular or transmembrane domains; pore lining, agonist/allosteric binding, and protein-protein interactions) was obtained from Brünger et al.35. Biophysical parameters were obtained from patch-clamp electrophysiology studies on Human Embryonic Kidney (HEK) 293 cells. The values were normalized with the respective wild-type receptor activation time (τr) and deactivation time (τd) constants.
Since our study is based on the idea that amino acid variants adjacent to or in proximity to functionally identified mutations in GABAA receptor subunits may exhibit similar patterns of electrophysiological changes in channel function, as observed in cases of these mutations, we explored the possibility of a relationship between structural, physicochemical, and biophysical parameters. The locations of the variants with respect to their distances from the membrane center and pore axis are given in Figure 5 and Figure 6. In this context, we also used the scores (Supplementary File 4: Supplementary Table S7) of AlphaMissense37; powered by the highly accurate protein structure prediction model AlphaFold264, which can utilize the basic amino acid sequence as input. AlphaMissense can provide clues for the structural aspects of single amino acid substitutions. The distribution of AlphaMissense scores for known (black) and predicted (red) variants with respect to variant position (amino acid position, distance to membrane center, and distance to pore axis) of GABAA receptor subunits (α1, β3, γ2 subunits encoded by GABRA1, GABRB3, GABRG2 genes respectively) is given in Figure 7.
Figure 7A-C shows the AlphaMissense score distribution across amino acid positions, Figure 7D-F shows the AlphaMissense score distribution over normalized distance from the pore axis, and Figure 7G-I shows the AlphaMissense score distribution over distance from the membrane center.The correlation analysis in Figure 7 indicated the difficulty of ascertaining an underlying relationship through structural properties to predict the outcome for newly identified variants. b2 subunit (encoded by GABRB2 gene) variants were included in the clustering and correlation sections to be able to conduct a wider analysis. However, only the variants of the α1 subunit encoded by GABRA1 (Figure 7A,D,G), β3 subunit encoded by GABRB3 (Figure 7B,E,H), and γ2 subunit encoded by GABRBG2 (Figure 7C,F,I) were included in the biophysical models, since the model focuses on the function of a hippocampal pyramidal neuron and the α1β3γ2 combination of GABAA receptor subunits is the most widespread combination in the hippocampus65. Similarly, any variants of α1, β3, or γ2 for which channel kinetics have not been studied in an α1β3γ2 GABAA receptor were also excluded from the simulations. There was a mild correlation (Supplementary Figure S1 and Supplementary Figure S2) between the AlphaMissense scores and biophysical parameters (normalized activation and deactivation times) derived from the effects of the GABAA receptor mutations (Supplementary File 4 and Supplementary Table S8) in the present analysis. This suggests that mutations predicted to be pathogenic (based on AlphaMissense scores) might also lead to measurable, potentially disruptive changes in receptor kinetics (e.g., activation and deactivation times). Nevertheless, the lack of correlation between the positional correlations in Figure 7 makes it difficult to utilize AlphaMissense scores for our assumption, which is based on the idea that adjacent amino acids will have similar consequences for biophysical characteristics.
The distributions of normalized distance to pore axis with respect to the activation and deactivation kinetics of the known variants are shown in Figure 8 and Figure 9. There is amild correlation for the γ2 subunit (Figure 8C), suggesting the possibility that our hypothesis, which is based on the assumption that adjacent amino acids will have similar consequences, may hold true in some regions, specifically in the proximity of the pore area of the receptor channel, the TM2 domain. This region is adjacent to our reference epileptogenic mutation (Figure 2 and Figure 4; γ2P302), making it a relatively good candidate for neural simulations. Based on this, a rough estimation of the effects of adjacent predicted mutations such as γ2A303T (Figure 2 and Figure 4) can be made. Our results presented here only consider the measurements on α1β3γ2; therefore, the variants assessed in our model were constrained to the variants given in Supplementary File 4:Supplementary Table S16.
Effect of mutations on GABAA receptor-mediated inhibition of CA1 pyramidal neuron firing
The effect of mutations on GABAA receptor-mediated inhibition is demonstrated on a multi-compartmental conductance-based CA1 pyramidal neuron model. The impact of GABAA receptor missense variants on the hippocampal pyramidal neuron function can be explored through the GABAergic shunting of apical inputs to the neuron, from the projections of CA3 and entorhinal cortex (EC) III pyramidal neurons. In other words, one way to simulate the activity of GABAA receptors is to assume a context in which the simulation represents realistic assumptions about the receptors' physiological significance, such as shunting inhibition, one of the mechanisms of GABAergic inhibition. The CA1 hippocampal pyramidal neurons, typically in their apical dendrites, have GABAA receptors at these zones, which are targeted by the projections from the neurons of the CA3 and EC III. This arrangement is thus suitable for the simulation. This research question requires an input design with varying delays and intensities. Therefore, three different glutamatergic synapses (GluS1/2/3) were placed on the distal apical, medial apical, and basal dendrites, as shown in Figure 10, and they were activated sequentially. For evaluating the impact of synaptic inputs, the constant current amplitude should remain below the minimum spike-triggering threshold (Iinj < Imin). The pyramidal neuron model with either wild-type or mutant GABAA receptor was initiated with a constant current injection of 0.85 nA at the soma. The GABAergic synapse was then placed at the soma. The presynaptic activity, mimicked by the spike generator, was initiated first at the distal apical dendrite. The synaptic inputs on the medial apical and basal dendrites were delayed by 25 ms and 50 ms, respectively. The GABAergic synapse was activated with a 40 ms delay. The GABAergic inhibition intensity was adjusted such that the whole spike train, except the first spike, is inhibited. Then, the impact of variants is explored in this setting by varying τrise, τdeactivation, and gGABAA .
The parameters for wild-type and mutant receptors were obtained from the collection described in protocol step 2.1.1 specifically for receptors composed of α1β3γ2, which is the most abundant subunit composition in hippocampal pyramidal neurons65. The parameter distribution is given in Supplementary File 4: Supplementary Table S16.
Each subunit mutation was tested on single, double, and triple glutamatergic synapse cases. In a simple approach, the impact of mutations can be evaluated over the firing rate and pattern. The ΔtISI averages and standard deviation can also be estimated to further gauge the changes in the firing pattern, where ΔtISI represents the change in interspike interval. The results for each case are given as firing rates, and ΔtISI (mean and standard deviation) in Supplementary File 4: Supplementary Table S17 and Supplementary Figure S3. The spike trains and voltage traces for the variants that altered the firing patterns are given in Figure 11, Figure 12, and Figure 13.
For single (GluS1) and triple (GluS1-2-3) glutamatergic synapse activation, the mutations that altered neuron response were only β3N110D and γ2K328M mutations. In the single glutamatergic input case, β3N110D led to impaired inhibition, and the firing pattern was locked on the glutamatergic spike train after the 4th presynaptic spike with a short delay (Figure 11). γ2K328M also impaired the inhibition, albeit only around the 5th presynaptic spike, and introduced a larger delay in postsynaptic spike compared to β3N110D (Figure 11). In the GluS1-2-3 activation case (Figure 13), the response was similar between β3N110D and γ2K328M mutations. Both mutants yielded a firing pattern where almost all the cumulated presynaptic spikes were detected and triggered a response. In both cases, neuron models fired with a spike pair in response to presynaptic activity.
The double glutamatergic synapse activation yielded distinct results compared to the other two settings (Figure 12). In this case, two mutations on the GABAA receptor b3 subunit (β3N110D and β3T288N) and two mutations on GABAA receptor γ2 subunit (γ2P302L and γ2K328M) impaired the GABAergic inhibition. The neuron model with γ2P302L mutant fired almost in synchrony with the GluS2, which was most likely a delayed response to the GluS1 with approximately the same delay of presynaptic spikes between GluS1-2. The β3T288N mutation yielded a similar result, with the distinction of the second spike still in synchrony with GluS2. The neuron model with the β3N110D mutant responded to almost every cumulated glutamatergic input, except for the first two presynaptic spikes of GluS1/2, which were introduced with a shorter ΔtISI. The firing pattern for γ2K328M was again like β3N110D, with the distinction of second and third presynaptic spikes being missed.
These results demonstrate the diverse effects of b3 subunit (encoded by GABRB3 gene) and γ2 subunit (encoded by GABRG2) mutations on the hippocampal pyramidal neuron activity. Interestingly, the mutations β3L170R, β3A305V, β3E180G, β3D120N, β3Y302C, and γ2R82Q did not yield any change in neural activity. The most severe impairment on inhibition was for β3N110D and γ2K328M, both of which had significantly lower τdeactivation and higher τrise. Our preliminary analysis also showed that changes in τrise or gGABAA alone are not enough to impair inhibition (data not shown). It can be argued that the mutations that lead to significantly decreased τdeactivation together with increased τrise leads to a more significant impairment in GABAergic inhibition.
In the case where all excitatory inputs must be inhibited, any mutation resulting in firing will yield abnormal and nonspecific neural responses, which have the potential to be exaggerated in a neural circuit composed of neurons with the same mutations. The balance of excitation/inhibition in a neural network can be significantly affected by the resulting impaired inhibitory feedback, which is a crucial component of any network activity.
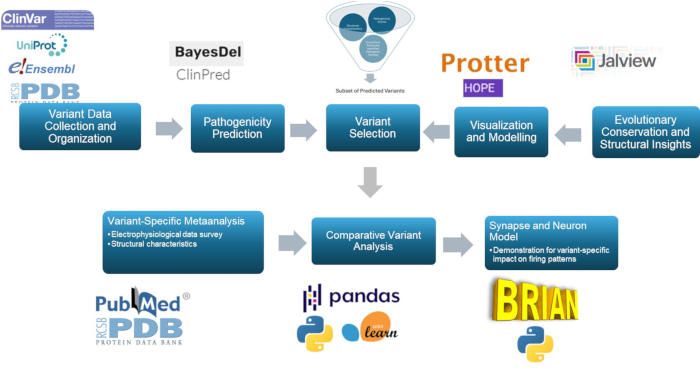
Figure 1: Overview of variant effect prediction and analysis for clinical and research purposes, with a specific focus on in silico analysis and neural response simulations. Please click here to view a larger version of this figure.
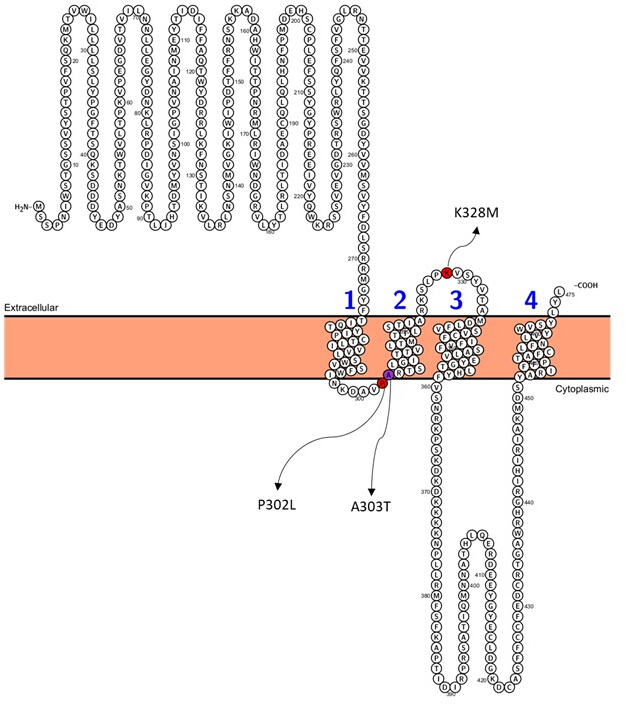
Figure 2: Position of γ2A303T and selected patient mutations;γ2P302L and γ2K328L used for neural simulations. Please click here to view a larger version of this figure.
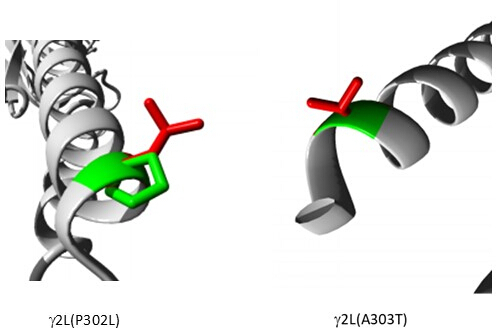
Figure 3: Comparative modeling of patient mutation γ2P302L and the adjacent variant γ2L(A303T) predicted as pathogenic. In both models, green represents wild-type and red represents mutant residues. Please click here to view a larger version of this figure.
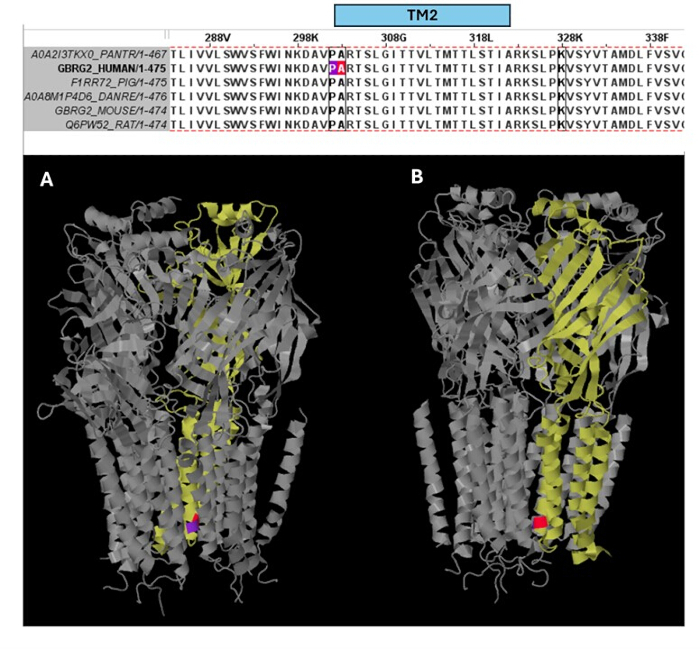
Figure 4: Multiple sequence alignment and structural insights. Top panel shows the evolutionary conservation of the residues in the position of the patient mutation (P302L) (purple) at the edge of the TM2 and of the pathogenic variant A303T (red color) in the beginning of the TM2 of the the γ2 subunit. Bottom panel shows the visualization of these conserved residues in the (A) three-dimensional GABAA receptor structure (7QNE), where the γ2 subunit (Chain C in the 7QNE) is shown as yellow and from different angles (A, B). Abbreviation: TM2 = second transmembrane domain. Please click here to view a larger version of this figure.
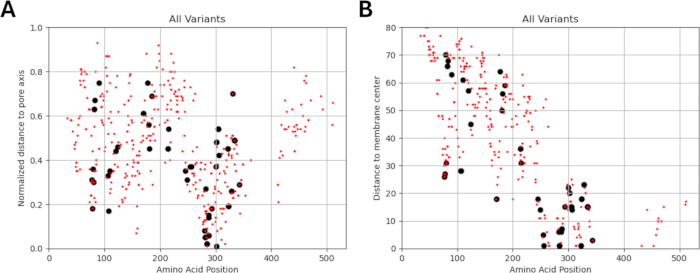
Figure 5: Localization of all included variants. The locations of known (black) and predicted (red) variants with respect to their (A) normalized distance from the pore axis and (B) distance from the membrane center are shown. Please click here to view a larger version of this figure.
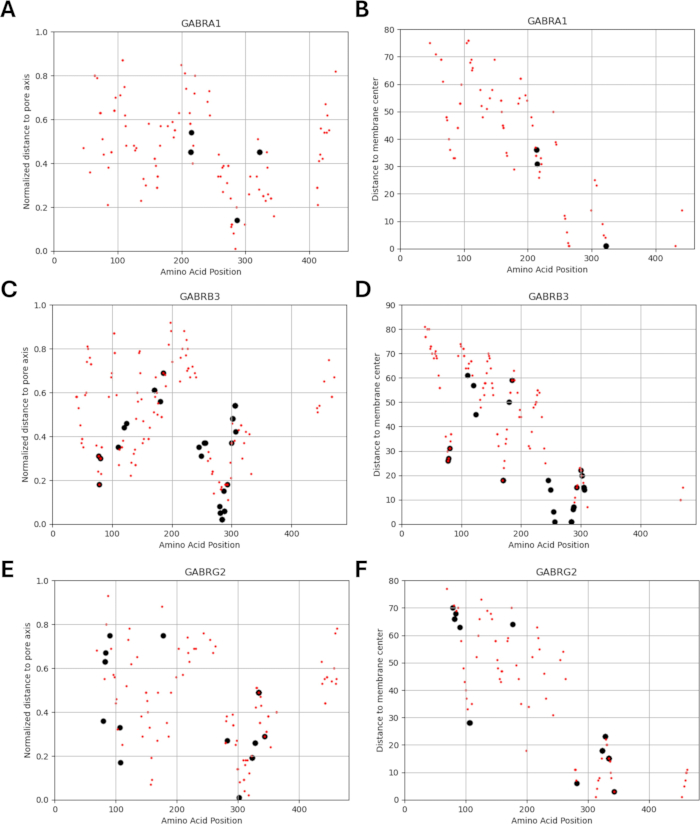
Figure 6: Localization of variants for each subunit. The locations of known (black) and predicted (red) variants with respect to their (A, C, E) normalized distance from the pore axis and (B, D, F) distance from the membrane center are shown. Please click here to view a larger version of this figure.
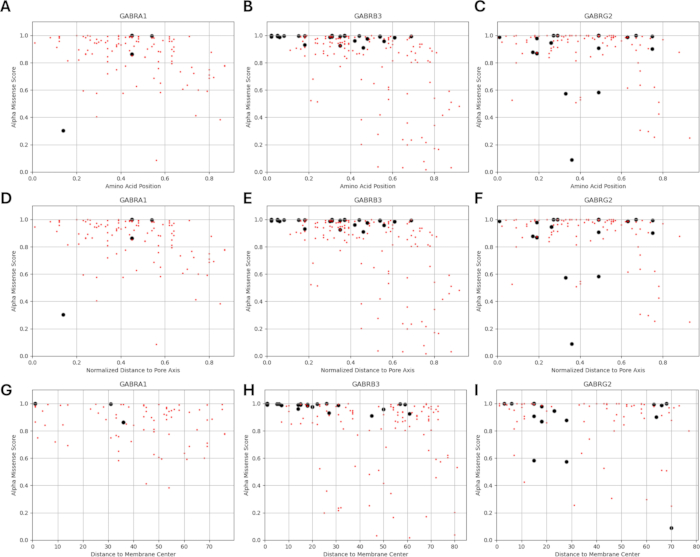
Figure 7: AlphaMissense score distribution over variant location. (A-C) AlphaMissense score distribution over amino acid position, (D-F) normalized distance from the pore axis, and (G-I) distance from the membrane center are given for known (black) and predicted (red) variants. Please click here to view a larger version of this figure.
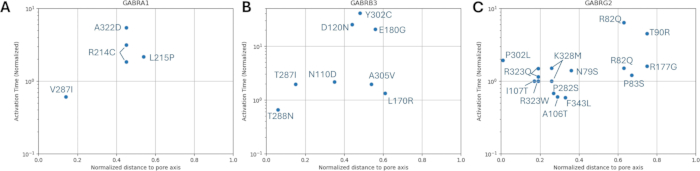
Figure 8: Normalized activation time of GABAA receptors with mutations in α1 (GABRA1) subunit, β3 (GABRB3) subunit, and γ2 (GABRG2) subunit. The experimentally obtained activation time constants with respect to normalized distance from the pore axis for each mutation on (A) α1, (B) β3, and (C) γ2 subunits are displayed. The values were normalized with the respective wild-type receptor activation time. Please click here to view a larger version of this figure.
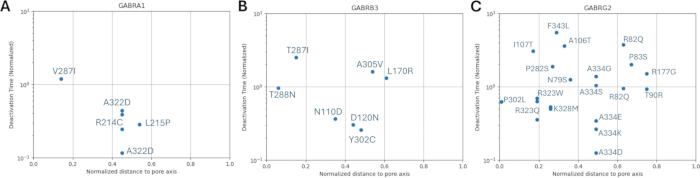
Figure 9: Normalized deactivation time of GABAA receptors with mutations in α1 (GABRA1) subunit of GABAA receptor, β3 (GABRB3) subunit of GABAA receptor, and γ2 (GABRG2) subunit. The experimentally obtained deactivation time constants with respect to normalized distance from the pore axis for each mutation on (A) α1, (B) β3, and (C) γ2 subunits are displayed. The values were normalized with the respective wild-type receptor deactivation time. Please click here to view a larger version of this figure.

Figure 10: CA1 pyramidal neuron model. The model neuron consists of (1) soma, (2) an apical dendrite with proximal, medial, and distal compartments, ending with two branches at lamina molecularis, (3) two symmetrically composed basal dendrites that branch into two sections after a short basal dendrite stem, and (4) an axon that starts with a conical axon hillock, followed by cylindrical axon initial segment, myelinated segments, and nodes of Ranvier, ending with a spherical axon terminal. The green triangles indicate the location of glutamatergic synapses, and the red triangle represents the GABAergic synapse located at the soma. Scale bar = 100 µm. Please click here to view a larger version of this figure.
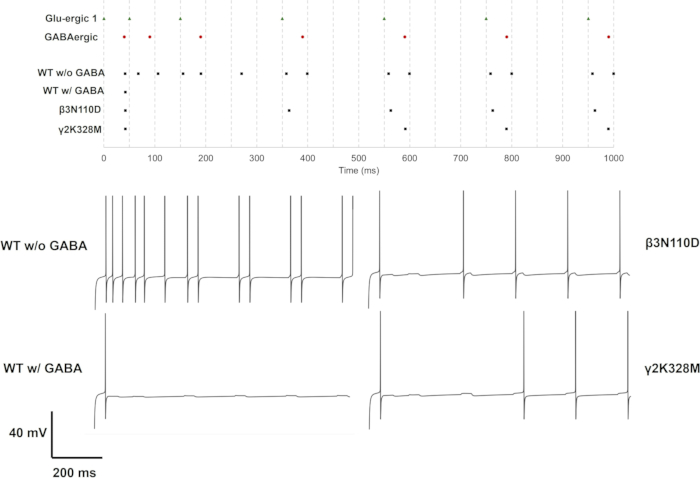
Figure 11: Firing pattern with only GluS1 activity. The spike trains for presynaptic neurons (GluS1 (green triangle) and GABAergic (red circle)) and the postsynaptic neurons with either wild-type or mutant GABAA receptors (black square) are given in the upper panel. Individual voltage traces for neurons with wild-type GABAA receptor with or without GABAergic inhibition and for neurons with mutant GABAA receptors with GABAergic inhibition are displayed in the lower panels. Please click here to view a larger version of this figure.

Figure 12: Firing pattern with only GluS1 and GluS2 activity. The spike trains for presynaptic neurons (GluS1/2 (green triangle) and GABAergic (red circle)) and the postsynaptic neurons with either wild-type or mutant GABAA receptors (black square) are given in the upper panel. Individual voltage traces for neurons with wild-type GABAA receptor with or without GABAergic inhibition and for neurons with mutant GABAA receptors with GABAergic inhibition are displayed in the lower panels. Please click here to view a larger version of this figure.
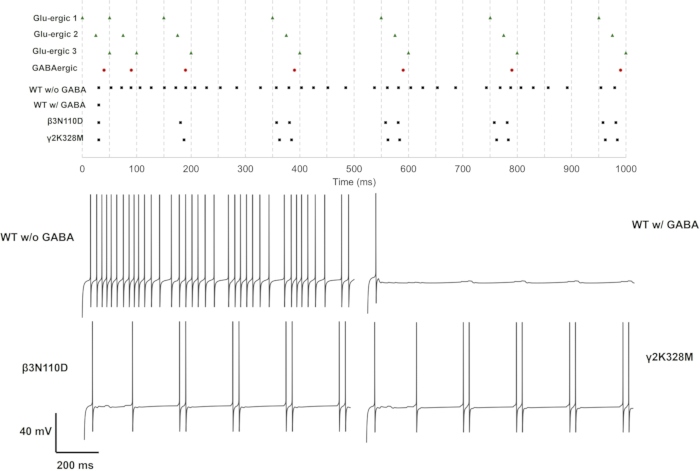
Figure 13: Firing pattern with only GluS1, GluS2 and GluS3 activity. The spike trains for presynaptic neurons (GluS1/2/3 (green triangle) and GABAergic (red circle) and the postsynaptic neurons with either wild-type or mutant GABAA receptors (black square) are given in the upper panel. Individual voltage traces for neurons with wild-type GABAA receptor with or without GABAergic inhibition and for neurons with mutant GABAA receptors with GABAergic inhibition are displayed in the lower panels. Please click here to view a larger version of this figure.
Supplementary Figure S1: Distribution of AlphaMissense scores and biophysical parameters (Normalized deactivation time; Normalized τd) of GABAA receptor subunit mutations selected in the present study. Also see Supplementary File 4: Supplementary Table 8. Please click here to download this figure.
Supplementary Figure S2: Distribution of AlphaMissense scores and biophysical parameters (Normalized activation time; Normalized τr) of GABAA receptor subunit mutations selected in the present study. Also see Supplementary File 4: Supplementary Table 8. Please click here to download this figure.
Supplementary Figure S3: The interspike intervals for neural response with wild-type and mutant GABAA receptors. The uppermost plot indicates the interspike time intervals for single glutamatergic input. The middle plot shows two, and the lowermost plot shows three glutamatergic synapses active simultaneously. Please click here to download this figure.
Supplementary File 1: The file "Data_GABAA.R" required to run in R for data formatting. Please click here to download this file.
Supplementary File 2: Equations used in the design of Conductance-based Model. Please click here to download this file.
Supplementary File 3: GABAAvar.py required to run in Brian2 for Neural Simulation. The file contains the Python codes for Brian2-based multicompartmental neuron model (function: CA1_Pyr), equations for conductance-based neuron and synaptic models (function: model_eqns, syn_eqns) and initial parameters (function: biophys_param, morpho_param, syn_param). Please click here to download this file.
Supplementary File 4: A zip folder containing all Supplementary Tables. Please click here to download this file.
Supplementary Table S1: Missense variants of unknown significance in the GABRG2 gene downloaded from ClinVar as .txt file and subsequently saved as "data.xlxs." Please click here to download this table.
Supplementary Table S2: Identifiers of the sequences used in the study, the reference transcript of the gene of interest (NCBI Ref. seq.) and other corresponding identifiers across different databases). Please click here to download this table.
Supplementary Table S3: Positions of the structural and functional regions. The positions of the specific regions of the γ2 subunit protein (NCBI Reference Sequence: NP_944494.1) encoded by the reference transcript (NM_198904.4) Please click here to download this table.
Supplementary Table S4: The content of the file "data1.xlxs," which represents ClinVar data of GABRG2 that includes only the columns: "GRCh38Chromosome", "GRCh38Location", "Name", "Protein change". Please click here to download this table.
Supplementary Table S5: The content of the "data1_output.xlsx" file that contains the required formatting of missense variants of GABRG2 data to be uploaded to the dbNSFP server for variant effect prediction. Please click here to download this table.
Supplementary Table S6: The content of the "data2.xlsx"file that contains the output from dbNSFP server for the variant effect prediction for unknown missense variants of GABRG2. Please click here to download this table.
Supplementary Table S7: AlphaMissense scores for GABAA receptor subunit variants. Please click here to download this table.
Supplementary Table S8: Biophysical characteristics of variants. Values for the biophysical parameters were obtained from previous studies with electrophysiology experiments. Variants are labeled with "S" (substitution) type, whereas the wild-type receptor parameters are given for each substitution and labeled with "C" (control). τd : Deactivation time constant, POpen : Open probability, gGABA: Receptor conductance, Imax: Maximum current, τr : Activation time constant. Please click here to download this table.
Supplementary Table S9: Physicochemical characteristics of variants. Previously identified variants with biophysical parameters are labeled with "S" (substitution) type, whereas the predicted variants are represented with "P". H: Change in hydrophobicity, VSC: Change in volume of the side chain, P1: Change in polarity, P2: Change in polarization, SASA: Change in solvent-accessible surface, NCISC: Change in net charge index. The values are obtained for each original amino acid and variant from Guo et al.36 and the change in each parameter is estimated as given. Please click here to download this table.
Supplementary Table S10: Structural characteristics of variants. Previously identified variants with biophysical parameters are labeled with "S" (substitution) type, whereas the predicted variants are represented with "P." The localization of the variant in a domain is represented with 1, else 0. All values are obtained from Brünger et al.35. Please click here to download this table.
Supplementary Table S11: Structural and biophysical parameter correlations for all known variants. Please click here to download this table.
Supplementary Table S12: Structural, physicochemical, and biophysical parameter correlations for known GABRA1 variants. Please click here to download this table.
Supplementary Table S13: Structural, physicochemical, and biophysical parameter correlations for known GABRB2 variants. Please click here to download this table.
Supplementary Table S14: Structural, physicochemical, and biophysical parameter correlations for known GABRB3 variants. Please click here to download this table.
Supplementary Table S15: Structural, physicochemical, and biophysical parameter correlations for known GABRG2 variants. Please click here to download this table.
Supplementary Table S16: Biophysical parameters for wild-type and mutant α1β3γ2 GABAA receptors. Please click here to download this table.
Supplementary Table S17: Firing rate and interspike intervals in response to single, double, or triple glutamatergic synapses with wild-type and mutant GABAA receptors. Please click here to download this table.
Discussion
By applying a combination of computational genetics, molecular modeling and neural simulations, the approach presented in this paper has the potential to improve the classification of GABAA receptor variants, offering valuable insights for both epilepsy research and clinical applications. A comprehensive analysis for the identification and prioritization of predicted pathogenic mutations is presented and extended into a framework that potentially bridges the gap between variant effects on protein and cellular phenotype. Evaluating the impact of epileptogenic GABAA receptor activity on hippocampal pyramidal neuron simulation allows the replication of in vitro phenotype associated with GABAA receptor dysfunction and demonstration of single neuron response alteration in the root of network dysfunction. Based on these simulations of neural responses generated by epileptogenic mutations, a rough estimation of the functional effects of structurally proximal predicted mutations was explored. The predictions on the effect of predicted mutations on channel kinetics require a thorough analysis with known variant sets. Comparative analyses, such as those presented in this paper, and neural activity simulations provide critical insights for the further generation and improvement of predictive models focusing on variant effect on the protein function and neural pathology. Additionally, our methodology can be used to select and prioritize the most pathogenic variants among the unknown variants to examine variant effects linked to GABAA receptor-related neurodevelopmental disorders. For instance, receptor subunits tagged with fluorescent probes66,67,68,69,70 and carrying the predicted mutations can be expressed in vitro to study their trafficking, cell surface expression, and neurophysiology. Besides, animal models such as C. elegans may be considered to validate the effects of predicted mutations. For instance, CRISPR-Cas9 gene editing has been used to generate a deletion of unc-49, a C. elegans GABAA receptor, thus generating homozygous epilepsy-associated mutations in unc-49 or subunits of the human GABAA receptor71.
In general, the classification of variants benefits from the use of multiple levels of computational evidence, as recommended by ACMG-AMP12. This approach strengthens the reliability of variant classification by integrating different predictive tools and data sources, ultimately enhancing the accuracy of clinical assessments and improving the overall decision-making process in genomic diagnostics. In our methodology, the utilization of ensemble predictors, which combine the predictions of multiple tools, thereby fulfilling the requirement for multiple lines of computational evidence and eliminating the need to use different tools separately is an advantage. This approach also overcomes the challenge of handling disparate outputs from individual tools, thus streamlining the prediction process and enhancing efficiency. Nevertheless, there is no guarantee regarding the predictive accuracy of gene-centric or variant-specific analyses. This leads to the conclusion that gene-centric or variant-specific predictions should be performed under specific conditions adjusted for the specific contexts and goals15,72,73,74. For clinical interventions, this would require the assessment of predictive accuracy of in silico tools for a specific gene or subset of genes in the context of a given disease often with individualized optimization75. However, the assessment of the predictive accuracy is often limited by the lack of a sufficient number of variants, which can affect the reliability of the accuracy assessment.
Different tools in the literature are available, and their accuracy is tested and validated in datasets14. However, these accuracy results based on large datasets are not necessarily reflected on the prediction of a few unknown variants for a given gene. In this context, accumulating literature suggests that ensemble predictors, which compile and compute the results of individual predictors, are known to perform better than concordance of individual predictors33,76,77,78 and thus, in the present study, we have chosen to use ensemble predictors, namely BayesDEL33 and ClinPred32 specifically for their superior performance32,34 BayesDEL was comparatively evaluated for 4,094 missense variants in clinically relevant genes, including genes encoding for transmembrane proteins such as voltage-gated sodium channel alpha subunit 5 (SCN5A), and showed superior performance33. In our variant effect prediction protocol, as a first step, we considered the consensus from two ensemble predictors (BayesDEL and ClinPred). AlphaMissense37, a deep learning model developed by Google DeepMind, is an extension of AlphaFold64,79, thus utilizing the power of high-accuracy protein structure prediction. When we compared the initial prediction results of ensemble models (BayesDEL and ClinPred as described in our protocol step 1.3) with the results of AlphaMissense, the predictions were partially in agreement with each other (Supplementary File 4: Supplementary Table S15) and did not fully align with the predictions of ensemble models (BayesDEL and ClinPred), which reached a consensus of pathogenic or disease-associated, shown as pink rows (Supplementary File 4: Supplementary Table S15). However, the unknown variants (L81F, A303T, and V329F) near the GABRG2 mutations R82Q, P302L, and K328M, which we used in our neuron model, and predicted as pathogenic both by ClinPred and BayesDEL, were also predicted as pathogenic by AlphaMissense as shown by yellow highlights (Supplementary File 4: Supplementary Table S15).
As AlphaMissense29 uses sequence and structural context prediction, in our study, we also wanted to see if there was any association between the AlphaMissense scores and GABAA receptor mutation locations based on their distances from the membrane center and pore axis. Our hypothesis is based on the idea that functional impact of amino acid variants adjacent or proximal to the functionally identified mutations of GABAA receptor subunits may show similar patterns of physicochemical changes in the channel function observed in the cases of mutations. A correlation between GABAA receptor subunit mutation positions and AlphaMissense scores would help us identify usable relationship to build a framework for our hypothesis allowing the prediction of the functional consequences of novel missense variants in GABAA receptor subunits. However, AlphaMissense scores were not predictive of changes in these biophysical parameters (Figure 7). It is important to note that the limited sample size in our analysis makes it difficult to draw definitive conclusions. However, our analysis found that the AlphaMissense scores did not correlate with the GABAA receptors structural parameters. The lack of a clear positional correlation (e.g., between mutations' positions and the AlphaMissense scores) challenges the validity of our assumption. If adjacent residues were truly having similar effects, we would expect to see a clearer correlation. Since this is not the case, it weakens the ability to use AlphaMissense scores as a reliable tool for testing our assumption.
Interestingly, in our study, we have found mild correlation between the distance of variant to pore axis and normalized channel activation time for the GABRG2 gene mutants. Thus, our preliminary assumption that adjacent amino acids will have similar consequences may hold true in some regions of the channel such as regions in the pore or at key sites involved in gating but may not be as clear-cut in other regions. The small dataset limits the ability to discern this variability, but future data or more detailed structural analysis could help refine this aspect of our hypothesis. Molecular dynamics simulations80 could serve as a powerful complementary approach to further investigate these preliminary findings especially in the context of comparative evaluation of two adjacent γ2 subunit mutations, namely the epileptogenic mutation γ2P302L40 and the proximal predicted mutation γ2A303T (rs1581439874), performed in our study. In the future, this approach could enable more accurate estimation of the effect of unknown variant on cellular phenotype, especially when integrated with the neural simulations presented in our study.
Additionally, it will be interesting to explore whether the structural and physicochemical properties of GABAA receptor subunits together with other features can be used to train powerful machine learning models for the functional prediction of novel variant effects on the channel, neuron, network and disease phenotype. With the advent of automated machine learning approaches, we have reached a point where doctors and wet lab scientists can also develop their own models in a more democratized environment81. Therefore, the integration of these technologies into clinical practice could potentially streamline the process, making personalized medicine more accessible and reducing the reliance on highly specialized expertise for functional variant analysis. In this context, our approach provides insights into the structural and functional dynamics of the receptor, potentially aiding in future studies for the functional prediction of variant effect.
Despite the current progress in protein structure prediction and the breakthrough represented by AlphaFold64, accurate prediction of the effect of mutations and protein function remains a challenge due to the lack of data required to train the model79. For the prediction of variant effect, AlphaMissense shows higher performance compared to a subset of predictive models but ensemble predictors BayesDEL25 and ClinPred24, which were used in our study, were not included in this comparison29. It is important to note that, in our study, the in silico tools BayesDEL, ClinPred, and AlphaMissense were employed for distinct purposes. The ensemble predictors, BayesDEL and ClinPred, were primarily used for pathogenicity prediction, whereas AlphaMissense was specifically used to explore the relationship between its scores and the known data for the impact of mutations in the γ2 subunit. Specifically, our hypothesis assumes that predicted pathogenic variants, particularly those located near or adjacent to functionally identified mutations in GABAA receptor subunits, will exhibit biophysical parameters similar to those observed in functionally characterized mutations. To investigate this, we chose AlphaMissense due to the fact that it is powered by the highly accurate AlphaFold264 model, which utilizes the basic peptide sequence to predict the consequences of single amino acid substitutions.
Consequently, a major limitation of our study is primarily driven by the limited availability of experimental data. For instance, our neuron model is based on the expression of the data derived from α1β3γ2 subunit combination of GABAA receptors, which inherently limits the mutations studied in the literature to the subunits expressed as part of this specific receptor combination. Additionally, we relied on electrophysiological data derived exclusively from the expression of these subunits in HEK cells, further narrowing the scope of available data in the literature. Our use of neural modeling to estimate the effects of unknown variants assumes that unknown variants (predicted as pathogenic in our workflow) located in close proximity to known mutations will exhibit similar patterns in channel kinetics parameters or physicochemical properties of mutation effects described in the literature. This assumption, coupled with the need for electrophysiological data for specific receptor assemblies in HEK293 cells, reduces the amount of experimental data available for modeling. As a result of these constraints, the available data allowed us to model only a limited number of variants in α1β3γ2 subunits. However, training the neuron model for different subunit assemblies such as α1β2γ2 subunit combinations, α1β2δ, or α4β3δ subunit combinations, which have subunit-specific cellular-, circuit-, and network-level implications, will likely show broader applicability to various epilepsy types and neurodevelopmental disorders. In the future, with the increase of available electrophysiological data and studies focused on mutations in well-defined receptor assemblies and specific cell types, it may become possible to enhance the generalizability and accuracy of our approach.
The multi-compartmental conductance-based neuron models provide a powerful tool to generate predictions for the functional significance of receptor variants of the single-neuron response. This tool enables flexible definitions of both cellular/synaptic parameters and their locations to test any specific question. Simple spike generators used in this protocol can be replaced with other neuron models to study microcircuit activity. The critical step of the protocol is also the most limiting step: the definition of any receptor variant in terms of altered channel kinetics. The required information is ideally provided by patch-clamp electrophysiology studies; however, the computational analysis of amino acid substitutions with predicted clinical significance and the comparisons with known substitutions can also provide some insight. Our study and described protocol incorporate the use of neural activity simulations not as a predictive tool, but rather as a tool to explore the effects of mutations to support a wider outlook on the consequences of altered biophysical characteristics of GABAA receptor on single-neuron activity. The dependence on experimental data in neural simulations is an important limitation in our approach, which might benefit from advanced molecular modelling to bridge the gap between structure and function.
In our protocol, certain steps should be critically evaluated. First, the choice of the predictive model used in the first part of our protocol may be critical. The selection of in silico tools depends on several factors including the generation of sufficient multiple lines of computational evidence for powerful prediction12. Ensemble predictors integrating the analysis of multiple predictive algorithms better fit with this recommendation thus preferred compared to individual predictors. There are different predictors, and their accuracy is usually tested in large datasets, which does not necessarily guarantee the accuracy of the predictive model used for unknown variant effects located in a specific gene. This is compensated by using two ensemble models, which collect and compute the prediction results from multiple predictors. Additionally, the cutoffs of the predictive models may be adjusted to increase specificity if the purpose is primarily to identify the most pathogenic variants. Setting appropriate cutoffs is important for balancing the trade-off between sensitivity and specificity and ensuring accurate classification of variants. In our study, we used the default cutoffs. We especially did not change the cutoff in favor of capturing variants that may be more likely to be pathogenic since this would decrease the scope of variants to be examined in multiple levels of our analysis as described in our workflow. It is also important to note that when choosing the structural data for the three-dimensional reconstruction of the protein of interest, there is a need for the preliminary review of the structural data in the literature. The structural examination of GABAA receptors has recently gained momentum with robust structural studies reporting the three-dimensional structure of different receptor assemblies in different conditions26,27,82,83,84,85. Given the availability of these data, in our study, we focused on the experimentally determined structures for the reconstruction of structural data. However, the prediction of AlphaFold64 may be favored for the analysis of other proteins that lack such experimentally determined data. For structural data derived from experimental studies, it is important to pay attention to the amino acid numbering. PDB numbering of amino acids may be different from UniProt numbering since the former may not include the signal peptide that is removed during protein maturation. Besides, chimeric proteins expressed in the experimental system may cause discrepancies. In this case, pairwise alignment of the sequence of interest with the sequence derived from the structural data will help preserve consistency. In the literature, the structural data for the γ2 subunit protein are based on different methods, including experimental methods such as electron microscopy (EM) and the high accuracy prediction method of AlphaFold. If the experimental method does not cover the desired sequence fully with high resolution, AlphaFold predictions may be used. In the present study, the structure 7QNE26 was chosen since it corresponds to cryogenic electron microscopic structure of human full-length synaptic α1β3γ2 GABAA receptor. This exactly represents the full-length subunit combination, which was the focus of the present study.
Additionally, for comparative analysis, the use of normalized parameters of channel kinetics should be preferred, as the values of these parameters might vary depending on the receptor subunit composition and experimental settings. For instance, τrise and τdeactivation should be determined over x-fold changes over the wild-type control values. In protocol step 2.5, for a low number of known variants, parametric or categorical correlation analysis and determination of rho and p values might be preferred. Ideally, methods such as principal component analysis are expected to yield more accurate relationships but require a higher number of samples.
The simulation environment can be altered. In this study, Brian2 was preferred for the following reasons: the spatialneuron class in Brian2 provides a valuable tool to simulate neural activity. Brian2 has a significant advantage in using differential equations to describe continuous dynamics and update statements for discrete events instead of relying on predefined "black-box" models, and this leads to excellent code readability and adaptability since every aspect of the model can be explicitly defined in a single Python script, with the model equations stated in mathematical notation and only a tiny amount of Brian-specific vocabulary used37. Because the model is explicitly described, all features are documented and can be found in the primary simulation description file, which eliminates the need for the previously relied-upon "black-box" models, as mentioned in studies by Stimberg et al.45,86.
The current neuron model utilizes modified Hodgkin-Huxley type conductances46 with Na+, K+ and leak current. This conductance-based model can be further expanded by the inclusion of several other channel types, such as Ca2+ channels. For synapse models, it is important to note that these parameters should be obtained for specific subunit compositions, and only the variants with parameters measured with these compositions should be assessed. In this study, the α1β3γ2 receptor composition was selected; therefore, only the variants of α1, β3, or γ2, with channel kinetics parameters measured on an α1β3γ2 GABAA receptor, were included.
The estimation of cellular biophysics involves segmenting the cell into multiple cylindrical compartments, each potentially possessing varying conductivity characteristics. Despite the irregularities in the dendrites of a neuron, they can be viewed as locally homogeneous strands. Such models help in understanding the complex structure and functioning of cells. The model design focuses on a simplified version of the actual morphology that can reflect these features.
The location and physicochemical characteristics of the altered amino acid determine the impact on the channel kinetics. For example, an amino acid substitution resulting in an amino acid with a larger side chain will decrease the conductance of the channel if this change occurs on an amino acid lining the pore of the channel. The substitutions might also affect the opening/closing of the channel. For simplicity in this model, the GABA binding kinetics are reduced only to a ratio of available receptors; however, more detailed models can be designed to include this interaction to study the possible impact of substitutions that alter ligand binding affinity.
In conclusion, the present study employs computational methods to predict pathogenic variants in the GABAA receptor subunits and qualitatively estimate the possible cellular phenotype based on the simulation of epilepsy associated mutations in the hippocampal pyramidal neuron model. To the best of our knowledge, this is the first protocol to explore the combined application of computational genetics, molecular modeling, and neural simulations to assess how genetic variants may contribute to GABAA receptor dysfunction across multiple levels of complexity, from DNA to protein function and neural behavior. This protocol can provide a basis for improving estimation of potentially deleterious variants and associated neural pathology in epilepsy. Additionally, it may be utilized in the study of other channelopathies yielding important insights into the underlying mechanism of relevant neurodevelopmental and network disorders. Based on this, by incorporating GABAA receptor's structural and physicochemical evaluations to examine the functional impact of variants and their integration into GABAA receptor's channel kinetics, a more accurate analysis can be developed in the future.
Disclosures
All authors declare that they have no conflicts of interest related to this work.
Acknowledgements
We thank Çağla Koca for her assistance with the construction of the model neuron.
Materials
| Name | Company | Catalog Number | Comments |
| Brian2 | Sorbonne Université, INSERM, CNRS, Institut de la Vision, France; Imperial College London, United Kingdom | 2.8.0.4 | Stimberg et al., 2019 (https://pypi.org/project/Brian2/ ) |
| dbNSFP server | Genos Bioinformatics LLC, USA | v3.0 | Liu et al., 2020 (http://database.liulab.science/dbNSFP) (https://sites.google.com/site/jpopgen/dbNSFP) |
| HOPE | Centre for Molecular and Biomolecular Informatics CMBI, Radboud University, Netherlands | 1.1.1 | Venselaar et al., 2010 (https://www3.cmbi.umcn.nl/hope/) |
| Jalview | University of Dundee, UK | JV2 | Waterhouse et al., 2009 (https://www.jalview.org/) |
| Jupyter Notebook | Project Jupyter, USA | https://jupyter.org/install | |
| Phyton | Python Software Foundation, USA | 3.13 | https://www.python.org/downloads/ |
| Protter | ETH Zurich, Switzerland | Version 1.0 | Omasits, et al., 2014 (https://wlab.ethz.ch/protter/start/) |
| R | The R Foundation for Statistical Computing, USA | R version 4.3.2 | https://www.r-project.org/ |
| RStudio | Posit software, PBC, USA | RStudio 2023.12.1+402 "Ocean Storm" Release | https://posit.co/downloads/ |
References
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved